 |
Originally from a small town in Dominican Republic, Maria Teresa Rodolis graduated from SUNY New Paltz with a BSc in Chemistry. At SUNY New Paltz, Maria received the New York State Chancellor’s award for her research in the synthesis and testing of monocyclic β-lactams. She was awarded a graduate research fellowship through the National Science Foundation to conduct her current PhD research at the University of Warwick. Maria enjoys travelling, salsa dancing, rock climbing, yoga, hiking and teaching children.
|
Interaction of Translocase MraY with the Antibacterial E Protein from Bacteriophage ΦX174
Due to the discovery of antibiotics in the 1920’s many scientists lost interest in bacteriophage as potential antimicrobial agents. However, with the emergence of multidrug-resistant bacteria, a considerable amount of research is focusing on finding alternative cures for bacterial infections1.
Bacteriophage are viruses that utilise bacteria to replicate. Lytic phage use host proteins to replicate its genes and produce viral proteins. Once all viral proteins and genetic material are assembled, lytic factors are prompted to destroy the cell wall releasing the virus and bursting the cell.
Lytic factor E, encoded by bacteriophage ΦX174, was found to inhibit peptidoglycan biosynthesis via inhibition of translocase MraY. MraY catalyses the transfer of UDP-MurNAc-pentapeptide from the cytoplasm on to the membrane bound undecaprenyl phosphate carrier, leading to the formation of lipid I2.
In the presence of epitope-tagged Emyc Protein, the activity of MraY was found to decrease by 75% with a staggering accumulation of the UDPMurNAc-pentapeptide precursor3. In addition, these inhibitory properties were abolished when MraY was solubilised4. This suggests that lytic E forms a rigid protein-protein interaction with MraY in the membrane.
Under the supervision of Professor Timothy Bugg, Maria Rodolis, is studying the peculiar interaction between MraY and Lytic E protein. A model for the possible transmembrane interactions between protein E’s transmembrane helix and MraY’s helix 9 has been developed (Figure 1).
This model elucidates the possible formation of aromatic binding and salt bridge electrostatic interaction between protein E and MraY. Phe-288 can form aromatic π-bonding with Trp-4 and Trp-7 of protein E, while Glu-286 can form a salt bridge with Arg-3 of protein E. Maria Rodolis has synthesised analogues of protein E and has tested their inhibitory properties on membrane-bound MraY utilising a fluorescence inhibition assay develop by Agnes Mihalyi.
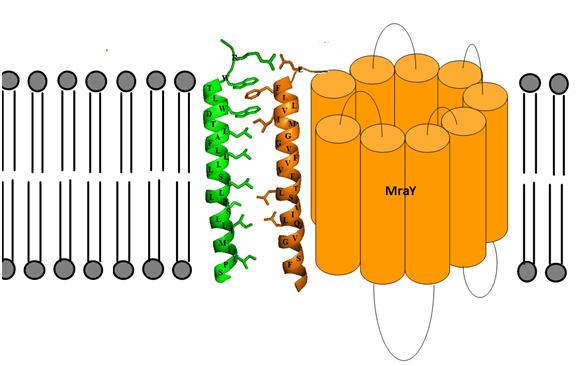
Figure 1: Predicted transmembrane interactions between Epep and MraY translocase.
References
1. Ghannad, M.S. & Mohammadi, A. Bacteriophage: Time to Re-Evaluate the Potential of Phage Therapy as a Promising Agent to Control Multidrug-Resistant Bacteria. Iran Journal of Basic Medical Sciences 15, 693-701 (2012).
2. Bouhss, A., Trunkfield, A.E., Bugg, T.D.H. & Mengin-Lecreulx, D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS microbiology reviews 32, 208-233 (2008).
3. Bernhardt, T.G., Struck, D.K. & Young, R. The lysis protein E of phi X174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. The Journal of biological chemistry 276, 6093-7 (2001).
4. Mendel, S., Holbourn, J.M., Schouten, J. a & Bugg, T.D.H. Interaction of the transmembrane domain of lysis protein E from bacteriophage phiX174 with bacterial translocase MraY and peptidyl-prolyl isomerase SlyD.  Microbiology 152, 2959-2967 (2006). Microbiology 152, 2959-2967 (2006).
|




