C3.0 Protein MSMS assignment software
This software program known as Cookson3.0 (C3.0) in honor of the mass spectrometrist it was co-developed with in order to replace him; Cookson Chiu (https://warwick.ac.uk/fac/sci/chemistry/research/oconnor/oconnorgroup/groupmembers/emeritusgroupmembers/cooksonchiu/), is a peptide + protein MSMS interpretation program, heavily inspired by the great work of Protein Prospector (https://prospector.ucsf.edu/prospector/mshome.htm) which many of us owe our start in tandem mass spectral interpretation to.
The program is written in LabView and compiled to a .exe file for easy distribution to any system to run without the full programming environment (only needing a driver, link when opened).
The Program has a "Prosite lite" design target of knowing your given sequence of biomolecule, then creating an extensive search space of fragments, Post Translational Modifications (PTM's), side chain loses (SCL's), and internal fragmentation channels. Thus it is focused on deep analysis of specific targets (such as new protein modifications/deep sequencing) rather than data base searching for peptide/protein identification.
The extensive array of fragments are then searched against the inputted peak lists, with options for various quality control checks, then outputting potential matches.
Cookson3.0 was written to provide the most extensive, comprehensive, and most importantly accurate calculations available for tandem MS data - which is critical for larger biomolecule species and more accurate data (such as that acquired via FT-ICR MS).
Dissatisfied with the previously available options/providers at the time of writing with respect to accuracy, fragment/modification options, elemental compositions, and output; Cookson3.0 was created. Though some programs - particularly the irreplaceable Protein Prospector, continue to be updated and provide new features and improvements to help the community since C3.0's conception.
Cookson3.0 boasts a wide variety of options for modifications + variations, custom amino acids+sequence changes, elemental composition output, custom elements, and export of the resulting assignments as a ready to publish table, mass error distribution plots+metrics, and output files to sequence cleavage map plotting.
Cookson3.0 is closed source (due to difficulty in distribution), available free on request, and will be sporadically updated/maintained. There are a great amount of features which can be added over time, many wish list items, and dreams, but a limited amount of hours to do so. Features can be added with great enough interest/need from anyone.
Please feel free to get in touch with questions/requests.
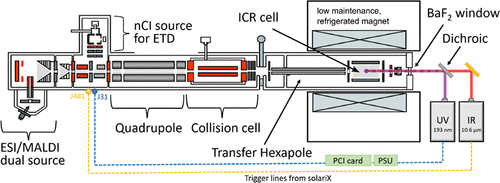
Cookson3.0 was recently releases along with various material for UVPD integration into a commercial FT-ICR MS platform. Please see the following publication reference:
- Alina Theisen, Christopher A. Wootton, Anisha Haris, Tomos E. Morgan, Yuko P. Y. Lam, Mark P. Barrow, and Peter B. O’Connor, Enhancing Biomolecule Analysis and 2DMS Experiments by Implementation of (Activated Ion) 193 nm UVPD on a FT-ICR Mass Spectrometer, Analytical Chemistry, 2022 94, 45, 15631–15638, https://doi.org/10.1021/acs.analchem.2c02354
This also houses a GitHub link to the repository for Cookson3.0, some example data, and will (in theory) be updated in future. If not; please ask directly.
