My Research
Analysing the kinetics of mRNP granule assembly and disassembly in Saccharomyces cerevisiae.
The control of translation competent mRNA numbers plays a vital role in the expression of proteins within the cell, and is highly regulated (Parker, 2012; Pérez-Ortín et al., 2013; Roy and Jacobson, 2013). One aspect of this control relies on the formation of microscopically visible non-membranous mRNP granules with varying composition, that act to control the level of free mRNAs by facilitating degradation, storage and re-entry into translation (Anderson and Kedersha, 2006; Brengues et al., 2005; Kedersha et al., 2005; Parker and Sheth, 2007). Each granule type contains sets of conserved protein orthologs (Buchan, 2014) and are conserved in eukaryotes, having been demonstrated in many model organisms including the budding yeast Saccharomyces cerevisiae (Hoyle et al., 2007). mRNP Granules not only have a role in general control of mRNA levels, but are also involved in stress-specific responses.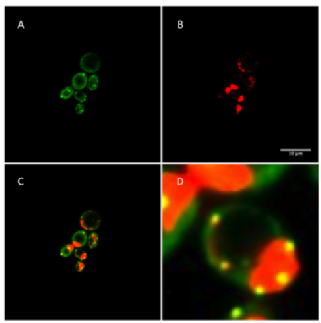
The 2 best studied mRNP granules types, P-bodies (PBs) and stress granules (SGs) exhibit distinct responses to a variety of cellular stresses including glucose deprivation, amino acid starvation, oxidative stress and heat shock (Aizer et al., 2014; Buchan et al., 2011; Iwaki and Izawa, 2012; Kramer et al., 2008). mRNP granules resembling to mammalian SGs have been also been identified as accumulating in a variety of neurodegenerative diseases, possibly due to the ‘prion’ domain in proteins that underpin their self assembly. High concentrations of these ‘prion’ domain proteins could form to stable aggregates and lead to cellular toxicity by sequestering mRNA and proteins within them (Buchan et al., 2011). An important question therefore is how these granules are cleared after the relief of stress, to avoid continual build up of the self-aggregating proteins. Traditionally this has been thought to occur by depletion of mRNA within the granules, either by exonuclease mediated decay, or re-entry into translation. The accumulation of PBs in mRNA decay pathway deletion mutations demonstrates the importance of mRNA levels in the turnover of these granules (Franks and Lykke-Andersen, 2008; Teixeira et al., 2005). One recent study highlights autophagy as a major route of clearance, demonstrating build up of stress granules in autophagy pathway protein mutants in both yeast and mammalian cells (Buchan et al., 2013).Although the mechanisms of assembly and disassembly have been partially uncovered, there remains a gap in knowledge as to how the cell decides on where to assembly granules, how large they become and how the occupancy of a single mRNA changes throughout the life-time of the granule. There appears to be a large degree of heterogeneity in granules size and number between cells, which could depend on any number of factors, from component concentration to endogenous signals such as position in the cell cycle (Franks and Lykke-Andersen, 2008). In this project, the aim will be to determine the cause of this heterogeneity in granule kinetics and more closely examine the sites of assembly and disassembly, focusing on the SGs and PBs in S. cerevisiae as a model for mRNP granules in higher eukaryotes.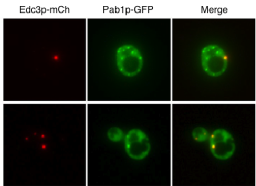
S. cerevisiae makes an excellent model organism as the mRNP granules that form within its cells are highly conserved with higher eukaryotes. It is however extremely genetically tractable and modification to proteins and mRNA are easily introduced in relatively short timescales (David and Siewers, 2014). Budding yeast can also harbour artificially genetic elements such as plasmids, allowing quick expression of modified proteins and nucleic acids. Exponential growth in liquid media means that protein complexes and components can be purified in large quantities and well understood mating mechanisms can facilitate high throughput genetic screening (Tong et al., 2001). There are also vast databases of yeast genetic and proteomic information available from high throughput studies via online resources. The ease of introducing genetic modifications to S. cerevisiae has also lead to wide use of fluorescent proteins, for microscopy and flow cytometry and more than 75% of known yeast ORFs have been tagged with GFP for high-throughput analysis (Huh et al., 2003). mRNAs can also be followed using fluorescent microscopy, using viral RNA sequences that fold into hairpin structures recognised by their cognate viral coat protein (Bertrand et al., 1998). These systems have been extensively used in yeast and other organisms to follow mRNA localisation in live cells such as that shown in figure 1. Budding yeast also have distinct morphology based on bud size and presence that make estimation of cell cycle phase possible using standard transmission light, phase contrast or DIC imaging, reducing the need for addition fluorophores and excitation light.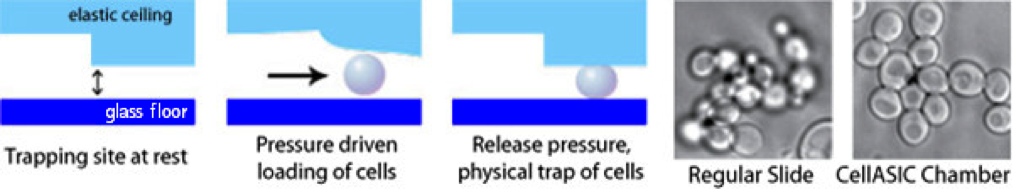
Both mRNP granules can be visualised by fusion of fluorescent proteins with their component proteins as shown in figure 2. Previous studies have attempted to address how individual granules, SGs or PBs form upon induction of the stress response using fluorescent microscopy but the time resolution of the imaging techniques used fails to capture this dynamic process(Buchan et al., 2008; Simpson et al., 2014). Here we propose to use advanced high speed imaging techniques, including but not limited to widefield deconvolution and spinning disk microscopy, to capture higher time resolution detail of the formation of mRNP granules. The use of a CellASIC microfluidic device (EMD Millipore, shown in figure 3) will allow media or chemical perfusion through live cells to image cellular stress in real time. Time-lapse imaging such as that shown in figure 4 will allow us to follow the lineages and phenotypes of each cell. Live cell tracking of the movement of these granules is also possible with the outlines techniques.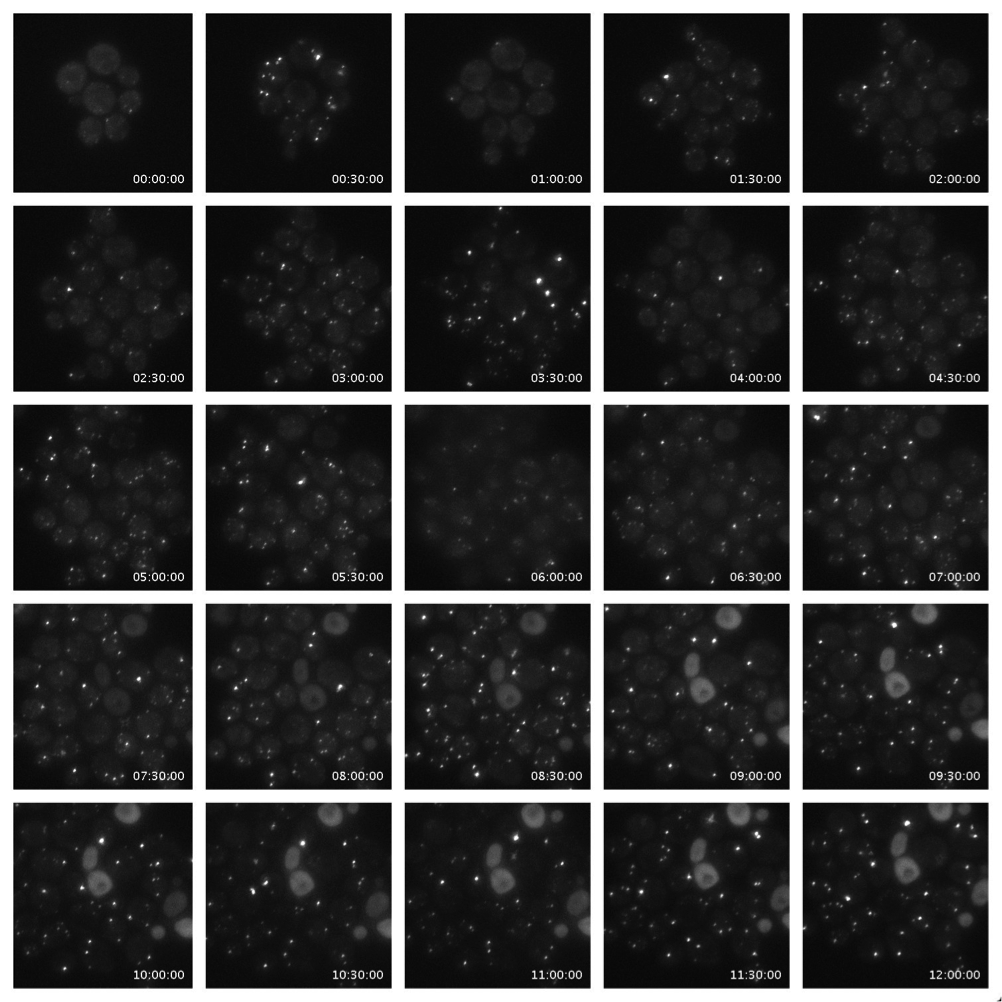
Using the outlined techniques we hope to advance the knowledge of how mRNP granules are assembled and disassembled. Microscopic analysis will give insight into how granules behave on a single cell level, what influences their formation and what impact they have on cell survival.
References:
Aizer, A., Kalo, A., and Kafri, P. (2014). Quantifying mRNA targeting to P bodies in living human cells reveals a dual role in mRNA decay and storage. J. Cell ….
Anderson, P., and Kedersha, N. (2006). RNA granules. J. Cell Biol. 172, 803–808.
Bertrand, E., Chartrand, P., Schaefer, M., Shenoy, S.M., Singer, R.H., and Long, R.M. (1998). Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445.
Brengues, M., Teixeira, D., and Parker, R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486–489.
Buchan, J. (2014). mRNP granules: Assembly , function , and connections with disease. RNA Biol. 1–12.
Buchan, J.R., Muhlrad, D., and Parker, R. (2008). P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183, 441–455.
Buchan, J.R., Yoon, J.-H., and Parker, R. (2011). Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 124, 228–239.
Buchan, J.R., Kolaitis, R.-M., Taylor, J.P., and Parker, R. (2013). Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474.
David, F., and Siewers, V. (2014). Advances in yeast genome engineering. FEMS Yeast Res. n/a–n/a.
Franks, T., and Lykke-Andersen, J. (2008). The Control of mRNA Decapping and P-Body Formation. Mol. Cell 605–615.
Hoyle, N.P., Castelli, L.M., Campbell, S.G., Holmes, L.E. a, and Ashe, M.P. (2007). Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179, 65–74.
Huh, W.-K., Falvo, J. V, Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S., and O’Shea, E.K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691.
Iwaki, A., and Izawa, S. (2012). Acidic stress induces the formation of P-bodies, but not stress granules, with mild attenuation of bulk translation in Saccharomyces cerevisiae. Biochem. J. 446, 225–233.
Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fritzler, M.J., Scheuner, D., Kaufman, R.J., Golan, D.E., and Anderson, P. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884.
Kramer, S., Queiroz, R., Ellis, L., Webb, H., Hoheisel, J.D., Clayton, C., and Carrington, M. (2008). Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J. Cell Sci. 121, 3002–3014.
Parker, R. (2012). RNA degradation in Saccharomyces cerevisae. Genetics 191, 671–702.
Parker, R., and Sheth, U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646.
Pérez-Ortín, J.E., Alepuz, P., Chávez, S., and Choder, M. (2013). Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 425, 3750–3775.
Roy, B., and Jacobson, A. (2013). The intimate relationships of mRNA decay and translation. Trends Genet. 29, 691–699.
Simpson, C.E., Lui, J., Kershaw, C.J., Sims, P.F.G., and Ashe, M.P. (2014). mRNA localization to P-bodies in yeast is bi-phasic with many mRNAs captured in a late Bfr1p-dependent wave. J. Cell Sci. 127, 1254–1262.
Teixeira, D., Sheth, U., Valencia-sanchez, M.A., Brengues, M., and Parker, R.O.Y. (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. 371–382.
Tong, a H., Evangelista, M., Parsons, a B., Xu, H., Bader, G.D., Pagé, N., Robinson, M., Raghibizadeh, S., Hogue, C.W., Bussey, H., et al. (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368.
Main Supervisor:
John McCarthy
John dot McCarthy at warwick dot ac dot uk
Co-supervisors:
Robert Cross
R dot A dot Cross at warwick dot ac dot uk
