Respiratory Syncytial Virus
RSV is the leading cause of severe viral pneumonia in infants and young children, and a major contributor to influenza-like-illness, including adults, throughout the world. No vaccine is available at present.
Unlike influenza, immunity to RSV is not long-lasting, which provides particular methodological challenges for modelling. Similarly, the role of multiple types (A and B), seasonality, wide and intermittently rapid genetic diversification and the relative strength of transmission in different contexts are a major focus of work. Finally, knowing how to integrate epidemiological, social behavioural and phylogenetic relatedness knowledge into design of interventions is a key task for our work on RSV.
Developing a vaccine to prevent disease in the most vulnerable age group – infants under 6 months of age – has been a focus for vaccinologists for over fifty years but poses real problems: young infants are essentially intolerant or unresponsive to RSV vaccines. This calls for exploration of the merits of a range of strategies that deliver vaccine to other groups that might directly or indirectly reduce RSV associated disease. Mathematical models lend themselves to this task. The potential targets include older infants, family contacts such as siblings and parents, school-going children or pregnant mothers (to boost passive antibody transfer to the infant). The benefit from such vaccination methods depends on features of the epidemiology of RSV that are not fully elucidated and are the subject of research project work being undertaken.
The following gives a flavour work we have been doing in this area with a view to developing decision tools for vaccine implementation.
Social contact studies – who contacts whom
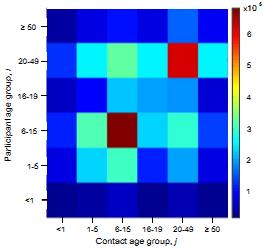 What are the underlying contact patterns that determine who transmits a respiratory virus to whom in the population? We have conducted personal diary based studies and evaluated the use of electronic tracking methods for determining contact patterns in a low income community in coastal Kenya. Diary based data from Kilifi, Kenya suggest an assortative nature to contacts as seen in developed country studies, as well as evidence of inter-generational high rates of contact (parents- children; school teacher-students) and different patterns between urban and rural settlements (of potential importance in a demographic transition.)
What are the underlying contact patterns that determine who transmits a respiratory virus to whom in the population? We have conducted personal diary based studies and evaluated the use of electronic tracking methods for determining contact patterns in a low income community in coastal Kenya. Diary based data from Kilifi, Kenya suggest an assortative nature to contacts as seen in developed country studies, as well as evidence of inter-generational high rates of contact (parents- children; school teacher-students) and different patterns between urban and rural settlements (of potential importance in a demographic transition.)
Contact rate surface (heat map right) expresses the mean number of contacts between an individual participant in age group i with an individual in age group j.
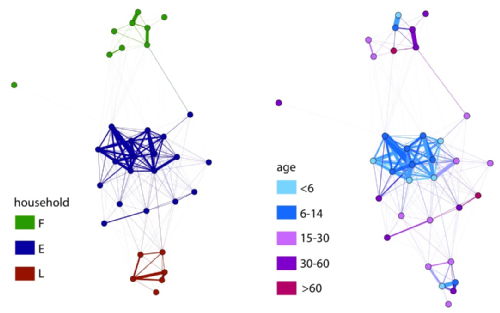 We are demonstrating the feasibility of using Radio Frequency Identification (RFID) tags in defining mixing networks in collaborative work with ISI Italy through the SocioPatterns project.
We are demonstrating the feasibility of using Radio Frequency Identification (RFID) tags in defining mixing networks in collaborative work with ISI Italy through the SocioPatterns project.
The network data (right) shows the importance of age group and household groupings in a local network: On the left, nodes are colour coded according to their household (E,F or L). On the right, nodes are colour coded according to their age. This work is due for expansion to saturate a larger population and to link to infection data.
Kiti MC, Kinyanjui TM, Koech DC, Munywoki PM, Medley GF, Nokes DJ. (2014) Quantifying age-related rates of social contact using diaries in a rural coastal population of Kenya. PLoS ONE 9(8): e104786.
Who infects the infant?
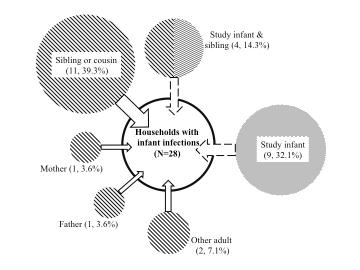 A key question is who introduces RSV into households and who subsequently infects the infant. We think we have a near definitive answer to the first of these – school age sibling or relatives presumably bringing infection back to the home from school or playing fields. We learned this from an intensive households surveillance study in a rural community in Kenya.
A key question is who introduces RSV into households and who subsequently infects the infant. We think we have a near definitive answer to the first of these – school age sibling or relatives presumably bringing infection back to the home from school or playing fields. We learned this from an intensive households surveillance study in a rural community in Kenya.
As for who actually infects the infant requires high resolution epidemiological and virus genetic relatedness data. As it stands sequencing the most variable gene of RSV (the G gene that encode s the attachment protein ) is not sufficiently diverse to resolve within-household contact tracking. We have therefore worked with the Sanger Instutute (Matt Cotton and Paul Kellam) to develop an RSV whole genome sequencing method, that looks to have promise simply froma numbers game perspective (ie while genome of ~15,000 Nt versus ~700 for ectodomain of G gene
Unknowns in the natural history of RSV
Surprisingly some basics on RSV infection and disease still require better quantification – to provide data suitable for modelling and predicting vaccination impact. In a birth cohort we have clearly shown that the risk of disease from RSV infection is independently and principally related to the age of the child (physiology – airway size). The infection history of the child (ie how many previous infection – an immunity issue) is of lesser importance.
We also show that RSV confers a short term partial immunity to reinfection (after each infection) in young children. The length of this immunity is of the order of 6 months that might be a fundamental determinant of the seasonal epidemic nature of RSV. However, in particular settings where there may be a very rapid build-up of susceptibles from birth or in-migration multiple epidemics per year could still be possible (roughly 6 month inter-epidemic period ) as seen in a short time series in the Dadaab refugee camp in NE Kenya in collaboration with CDC Kenya.
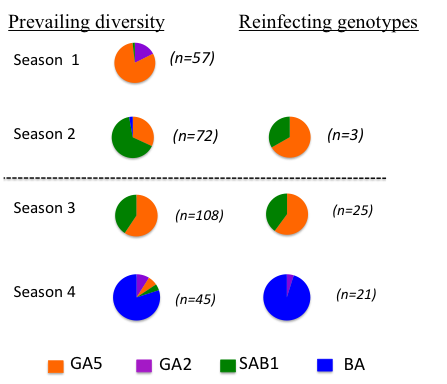 Are reinfections (which we know to be frequent) the result of loss of immunity or the result of evasion of the host immunity – ie evasion of strain specific immunity. The latter seems a possibility since the attachment protein G gene shows marked positive (immune) driven diversity and is a known neutralising antibody target. Counter to this, the F fusion protein, also on the surface of the virus shows relatively little diversity although is also a neutralisation antibody target. To tease this apart we sequenced the G gene of over 50 infection-reinfection paired viruses from a birth cohort.
Are reinfections (which we know to be frequent) the result of loss of immunity or the result of evasion of the host immunity – ie evasion of strain specific immunity. The latter seems a possibility since the attachment protein G gene shows marked positive (immune) driven diversity and is a known neutralising antibody target. Counter to this, the F fusion protein, also on the surface of the virus shows relatively little diversity although is also a neutralisation antibody target. To tease this apart we sequenced the G gene of over 50 infection-reinfection paired viruses from a birth cohort.
The majority of all reinfections were with distinct viruses – at the Group level (A or B), genotype within Group. This is suggestive of some form of strain specific immunity at the individual level.
However, the reinfecting strains were markedly in proportion to the distribution of circulating strains. The figure (right) compares, by epidemic, the genotype distribution in the general population with the reinfecting genotypes. Note that the majority of the reinfections were seen in the last two seasons.
The genetic relatedness of infecting and reinfecting strains of RSV
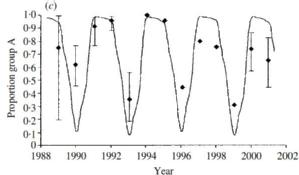 RSV occurs as two main circulating groups which show remarkable seemingly competitive sequential dominance. In the UK the pattern is predominance of group A for two epidemic seasons followed by group B dominance ie AAB cycling over each successive 3 year period (right). In Finland it is two epidemics (major-minor) of the same group, separed by a longer inter-epidemic period before two epidemics of the alternative group ie AaBa. The dynamics can be shown from models to be resultant from sequential development of population level group specific immunity [White et al] together with a slight difference in group specific transmissibilty (ie differential reproduction ratios: R0A>R0B
RSV occurs as two main circulating groups which show remarkable seemingly competitive sequential dominance. In the UK the pattern is predominance of group A for two epidemic seasons followed by group B dominance ie AAB cycling over each successive 3 year period (right). In Finland it is two epidemics (major-minor) of the same group, separed by a longer inter-epidemic period before two epidemics of the alternative group ie AaBa. The dynamics can be shown from models to be resultant from sequential development of population level group specific immunity [White et al] together with a slight difference in group specific transmissibilty (ie differential reproduction ratios: R0A>R0B
Modelling Transmission Dynamics of RSV
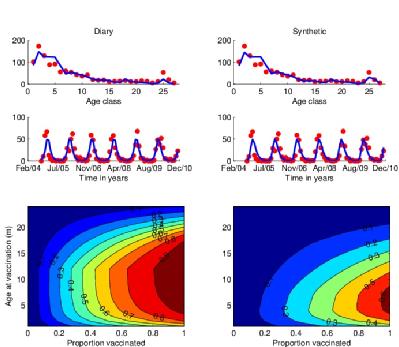 Using a realistic age-structured model of RSV we incorporate data from field and lab studies to explore different immunisation strategies. In the figure shown [Kinyanjui et al, in prep] we look at the merits of various ages at vaccination in the first year of life on the occurrence of severe RSV disease. As expected the optimal strategy is a compromise between vaccinating too early and resultant vaccine failure in those with maternally-derived immunity and vaccinating too late due to natural infection. However, the ‘window’ is quite wide and suggests that delaying vaccination into the second half of infancy might be highly beneficial through both direct protection of those vaccinated and indirect protection from reducing the circulation of viruses in individuals that would otherwise have contacted and infected the youngest infants. This results is highly dependent upon the contact structure of the population (determined from contact diary data (see above) or a household occupancy synthetic contact matrix), and understanding of the natural history of RSV infection in the host and epidemiology in the community.
Using a realistic age-structured model of RSV we incorporate data from field and lab studies to explore different immunisation strategies. In the figure shown [Kinyanjui et al, in prep] we look at the merits of various ages at vaccination in the first year of life on the occurrence of severe RSV disease. As expected the optimal strategy is a compromise between vaccinating too early and resultant vaccine failure in those with maternally-derived immunity and vaccinating too late due to natural infection. However, the ‘window’ is quite wide and suggests that delaying vaccination into the second half of infancy might be highly beneficial through both direct protection of those vaccinated and indirect protection from reducing the circulation of viruses in individuals that would otherwise have contacted and infected the youngest infants. This results is highly dependent upon the contact structure of the population (determined from contact diary data (see above) or a household occupancy synthetic contact matrix), and understanding of the natural history of RSV infection in the host and epidemiology in the community.
Funded by: Wellcome Trust, EPSRC
SBIDER people involved
James Nokes
Matt Keeling
Sam Brand
Additional Collaborators
Graham Medley
Thomas House
Tim Kinyanjui
KEMRI-Wellcome Trust Research Programme, Kenya
