Research interests
Over the last 25 years, the discovery of novel antibiotics has declined dramatically, and, in the past 2 years only 1 antibacterial agent was approved for clinical use in the US.[1] This would not be so alarming if the incidence of drug-resistant bacterial strains had stagnated, but these “Superbugs” have been proliferating rapidly.[1] With the antibiotic pipeline running dry, and with the pharmaceutical industries reluctance to invest in anti-infective discovery, novel strategies urgently need to be developed and applied to drug discovery.
Since the discovery of penicillin in 1928, the main source of antibiotics has been micro-organisms. More than 70% of commercially available antibiotics are produced by Streptomyces bacteria.[2] Following the sequencing of the entire genome of Streptomyces coelicolor A3(2), which is widely accepted as the model actinomycete, an unexpectedly large number of antibiotic-like gene clusters were found to be encoded within Streptomyces genomes.[3] Many of these so called cryptic gene clusters (cryptic because their products are not known) were predicted to encode for the biosynthesis of novel bioactive natural products. Such genome mining has resulted in the development of new strategies for isolating and characterising the metabolic products of these previously unknown gene clusters.[4]
Under laboratory culture conditions, these cryptic biosynthetic gene clusters are often not expressed. Consequently, new approaches are needed to access this untapped biosynthetic potential. The production of several Streptomyces secondary metabolites is triggered by gamma-butyrolactone (GBL) inducer molecules, the most characterised of which is A-factor (Fig. 1) made by Streptomyces griseus.[5] In the model streptomycete S. coelicolor A3(2), GBLs (S. coelicolor butyrolactones or SCBs, Fig. 1) directly regulate the production of a polyketide antibiotic of unknown structure.[6]
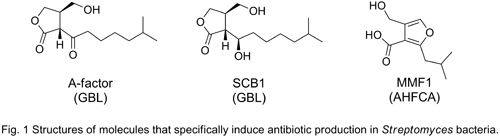
Through a genome mining approach, I have recently discovered a novel structural class of inducer molecules (2-alkyl-4-hydroxymethylfuran-3-carboxylic acids or AHFCAs, exemplified by MMF1, Fig. 1) made by S. coelicolor A3(2).[7] MMF1 specifically induces the production of methylenomycin A, one of the several antibiotics known to be made by this bacterium. Comparative genomics and a literature survey have shown the likely prevalence of AHFCAs in other bacteria. MMF1 is thought to interact with, and change the characteristics of, the DNA-binding proteins MmyR and MmfR. These two paralogous proteins have been shown to act as transcriptional repressors of the methylenomycin biosynthetic genes by binding to specific DNA sequences (AHFCA Responsive Elements or AREs).[8]
References: [1] Infectious Diseases Society of America, 2010, “Bad Bugs, No Drugs“; [2] D.A. Hopwood, "Streptomyces in Nature and Medecine: The Antibiotic Makers" 2007, New York, Oxford University Press; [3] S.D. Bentley, et al. Nature 2002, 417, 141; [4] C. Corre and G.L. Challis ChemBiol 2007, 14, 7; [5] M.J. Bibb Curr Opin Microbiol 2005, 8, 208; [6] K. Pawlik Arch Microbiol 2007, 187, 87; [7] C. Corre, et al. Proc Natl Acad Sci USA 2008, 105, 17510; [8] S. O’Rourke, et al. Mol Microbiol 2009, 71, 763
