Spatial Dynamics
Computationally efficient methods
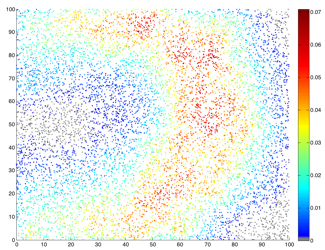 Example of spatial simulation of 10,000 individuals showing the probabilty of infection at peak of an epidemic. Example of spatial simulation of 10,000 individuals showing the probabilty of infection at peak of an epidemic. |
Simulation provides a flexible, and widely applicable, approach to understanding epidemic dynamics in the presence of randomness and uncertainty. Ultimately, the goal is to construct models where simulation has predictive power, at least in terms of reproducing an underlying distribution of random outcomes. Wide-scale repetition for large systems, such as repeated simulation of an epidemic spreading amongst the UK Farms, can be extremely computationally costly. Thus, there is strong incentive to develop faster, more computationally efficient, simulation methods. This is especially true for parameter inference, where many millions of simulations may be required. For spatial epidemics it is possible to use grid-based decompositions and transformation techniques to greatly increase the efficiency of drawing the next random event, and henceforth reduces the simulation time per iterate.
Incorporating human movement patterns





The above maps show the spread of a short-generation pathogen from a small number of initial cases in London. Spread throughout most of the UK has occurred within 30 days highlighting the need for rapid implementation of controls. Results are obtained by simulating the "meta-population" dynamics of infection within 10,000 wards in the UK; the spatial spread of infection is achieved using the work-place locations recorded in the 2001 census.
A wider spectrum of human movement behaviour can be captured by including information on non-work travel, for purposes such as shopping, study and irregular or unpredictable journeys. These movements can increase the rate at which diseases spread spatially. They can also play an important role in the dynamics of infections that are contracted from certain environmental sources. The figure below shows a fine scale spatial subdivision of a Welsh town, identifying particular shopping locations.
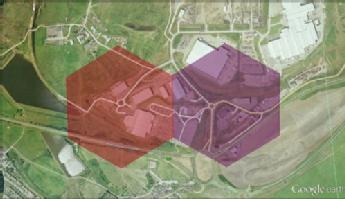
We are currently using this technology to investigate Legionella outbreaks, to understand how the predicted pattern of human movements can be used to inform the likely sources of infection. This approach could be particularly useful as a rapid means of focusing detection work, rather than relying on lengthy patient interviews (which may be complicated by memory-loss in Legionella patients).
Spatial ecology
A variety of plant species self-organise into large-scale structures such as bands and patches. How can we relate these spatial patterns to their underlying ecology? High-resolution data in the form of aerial and satellite photography provides a way of estimating important ecological processes (such as persistence and risk of extinction) from single snapshots in time. A variety of statisitical and mathematical techniques can be used to infer biological parameters from the spatial data.
One such species vital to our understanding is Seagrass. According to some estimates, Seagrass is one of the most ecologically and economically important basal species in coastal marine ecosystems. However, despite its relative importance, many aspects of its growth, population dynamics and spatial distribution are poorly understood. Seagrass meadows have extremely high primary and secondary productivity and support a great abundance and biodiversity of fish and invertibrates. In additon, the life cycles of some commercially important species has been linked to Seagrass. Yet Seagrass faces many threats including disease, pollution and the impacts of climate change.
Data provided by Natural England from Seagrass around the Scily Isles is being used to develop these techniques. In particular understanding how the fractal dimension of patch clusters relate to their ecology will be used to create cutting-edge ways of characterising risk of extinction useful to field ecologists.
Funded by EPSRC, Leverhulme, Wellcome.
WIDER people involved:
Matt Keeling
Leon Danon
Penny Hancock
Sam Brand
External collaborators:
Mark Temple (NHS Wales)
Ian Hall (HPE Porton)
