Cell division projects: modelling kinetochore dynamics, congression dynamics
I use a combination of mathematical modeling, model analysis and Bayesian inference techniques to develop an understanding of how chromosomes move and respond to forces during cell division. This is collaborative work with Andrew McAinsh (Warwick Medical School) who uses the latest microscopy techniques, including light sheet, to image chromosome dynamics during cell division. I currently have 4 projects on kinetochore (chromosome) dynamics, each discussed below.
- Chromosome dynamics in human oocytes. Funded by Wellcome Trust. Open position. Advertised.
- Chomosome congression dynamics. Funded by the BBSRC, 2018-2022. PDRAs Jonathan Harrison, Onur Sen (experimental component WMS).
- Intrakinetochore dynamics and force dependence. Funded by the Leverhulme Trust, 2018-2021. PDRAs Peter Embacher, Emanuele Roscioli (experimental component WMS).
- Tension-clock model analysis and inference. Previously funded by the BBSRC.
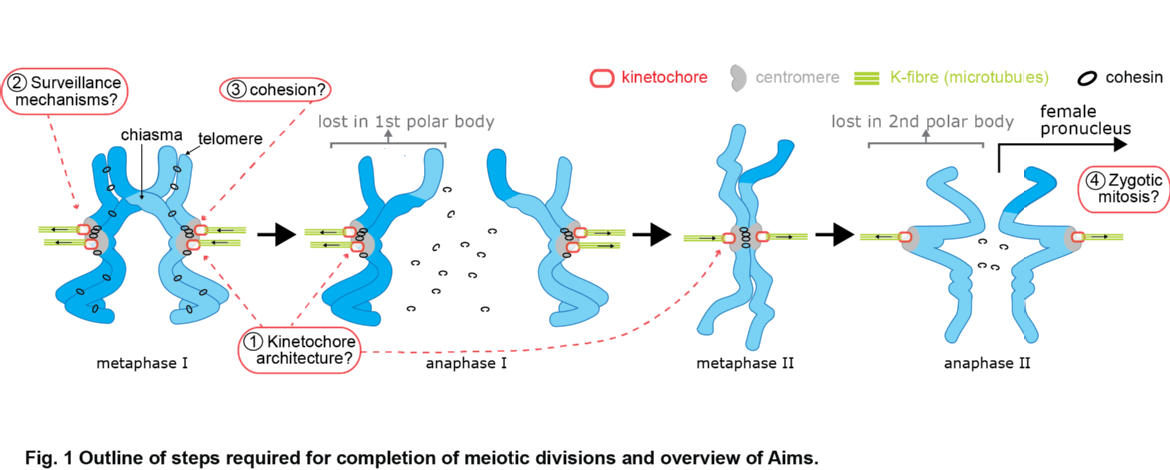
Chromosome dynamics in human oocytes (Wellcome Trust Collaborative grant (Warwick-Edinburgh), 2019-2024, open PDRA position).
How the human oocyte develops and the processes that organise the chromosomes from an initial cell with 92 paired recently duplicated chromosomes (1/2 from mother, 1/2 from father) to a cell with 23 (recombined) chromosomes is poorly understood. This is a complex mechanical process that is in fact highly error prone. This project uses a range of mathematical, computational and statistical techniques to analyse a range of high-end imaging data on human oocytes. The aim is to develop a quantitative understanding of how chromosome dynamic history can lead to errors, what predisposes a cell to make errors and whether errors can be corrected.
We work with experimentalists that will generate a large heterogeneous data set that is being used to determine the covariates of errors.
Congression dynamics (BBSRC, 2018-2022, PDRA Jonathan Harrison).
During cell division, the genome packaged in multiple chromosomes is duplicated. Ensuring each daughter cell then gets one, and only one copy of each chromosome is a mechanical process. In brief, duplicated chromosomes are held together (called sister chromatids), and dynamic microtubules generated from two spindle poles at opposite ends of the cell 'fish' for those chromatids. The desired configuration is to capture the sisters from opposite spindle poles (so called biorientation), the paired chromatids can then be positioned along the cell equator in a process called chromosome congression. However, since attachment is a random process, incorrect attachments occur which then go through rounds of 'error correction'. The range of reported states is illustrated in Figure 1. Once all chromatid pairs are aligned, anaphase occurs where the sisters are separated and pulled towards opposite spindle poles, the cell then breaks into two in a process called cytokinesis.
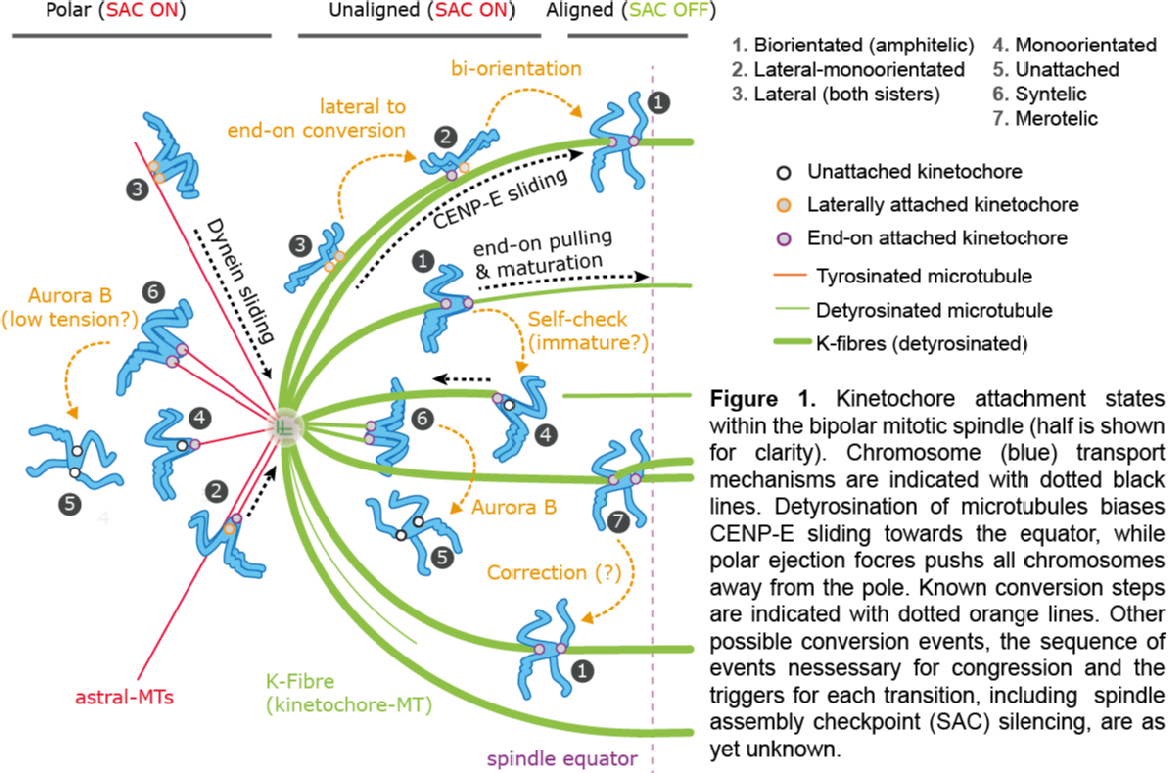
How microtubule attachments are monitored (thereby triggering or inhibiting error correction) is believed to occur through force sensing (tension between the sisters), but the details of this process remain to be established. We use sophisticated Bayesian techniques to fit mechanistic models to 3D trajectories (generated in Andrew McAinsh lab), using an iterative cycle to improve the models. This methodology firstly provides a means to automatically annotate trajectories into the various attachment states, and secondly provides estimates of key parameters and processes to aid interpretation of the congression process.
Intrakinetochore dynamics and force dependence. (Leverhulme Trust, 2018-2021, PDRAs Peter Embacher).
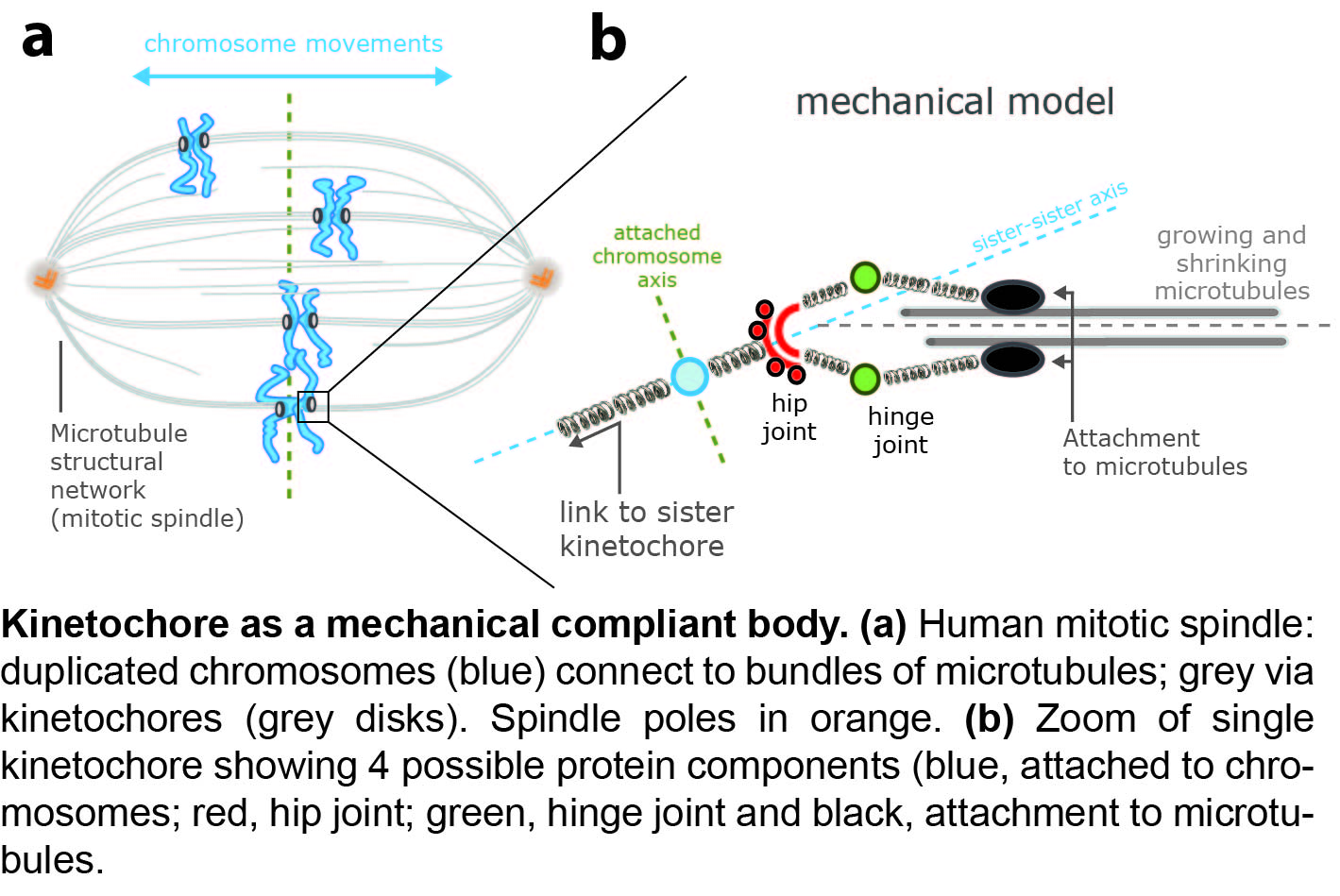
Microtubules attach to chromosomes via a protein complex called the kinetochore. This comprises 100+ different proteins and is beleived to be a force sensor and a force broker, regulating the state of attached microtubules and controlling chromosome movements. In collaboration with Andrew McAinsh, we are tracking the relative position and movements of the constituent proteins during chromosome oscillations in metaphase of the cell division cycle. Distances within the kinetochore are small, requiring sub-pixel resolution methods and analysis techniques to remove biases that occur when working with Euclidean distances. We discovered a new degree of freedom of the kinetochore, kinetochore axis swivel, eLife 2016, demonstrating that the kinetochore primary axis can swivel relative to the sister-sister axis, Figure 2. By extending this to a repertoire of 10+ flourescent tags we will construct a dynamic composite body model of the kinetochore to develop an understanding of what the compliance degrees of freedom are within the kinetochore, and thus how force and torques transmit through the kinetochore from microtubules to chromosomes.
Tension-clock model analysis and inference.
By reverse engineering chromosome dynamics during metaphase oscillations, we developed a new tension sensing model of chromsome directional switching, PLoS Comp Biology 2015, eLife 2015. I am analysis the oscillation dynamics of the deterministic and stochastic tension-clock model.