ePortfolio of Jie Zhang
2st Year post-graduate research student, School of Life Sciences
Supervisor: Prof. Daniel Hebenstreit
Significance of research
Transcription is a highly fluctuating process subject to stochastic variation, which produces great variations in the numbers of mRNAs that are found in individual, otherwise identical cells. This transcriptional cell-to-cell variability, often referred to as transcriptional noise, is incompletely understood, in particular in mammalian systems, and poses a problem for synthetic biology endeavors by making complex synthetic genetic networks unpredictable and unreliable. It is further implicated in causes of several diseases, including cancer and autoimmunity. There is currently limited understanding in the causes for transcriptional fluctuations but a major factor is believed to be ‘polymerase pausing’. This phenomenon refers to the apparent stopping of RNA polymerase II after transcriptional initiation. It is my major goal of this project to, for the first time, measure precise polymerase pausing times in transcriptome-wide fashion. To this end, I will modify the ‘PRO-seq’ assay and apply it to a mammalian cell line. I will then analyse the resulting data and test how it relates to mRNA expression levels and their single cell variation. For this I will include other, existing datasets based on bulk and single-cell RNA-seq. Eventually, my project will yield a much better understanding of the mechanism underlying transcriptional dynamics.
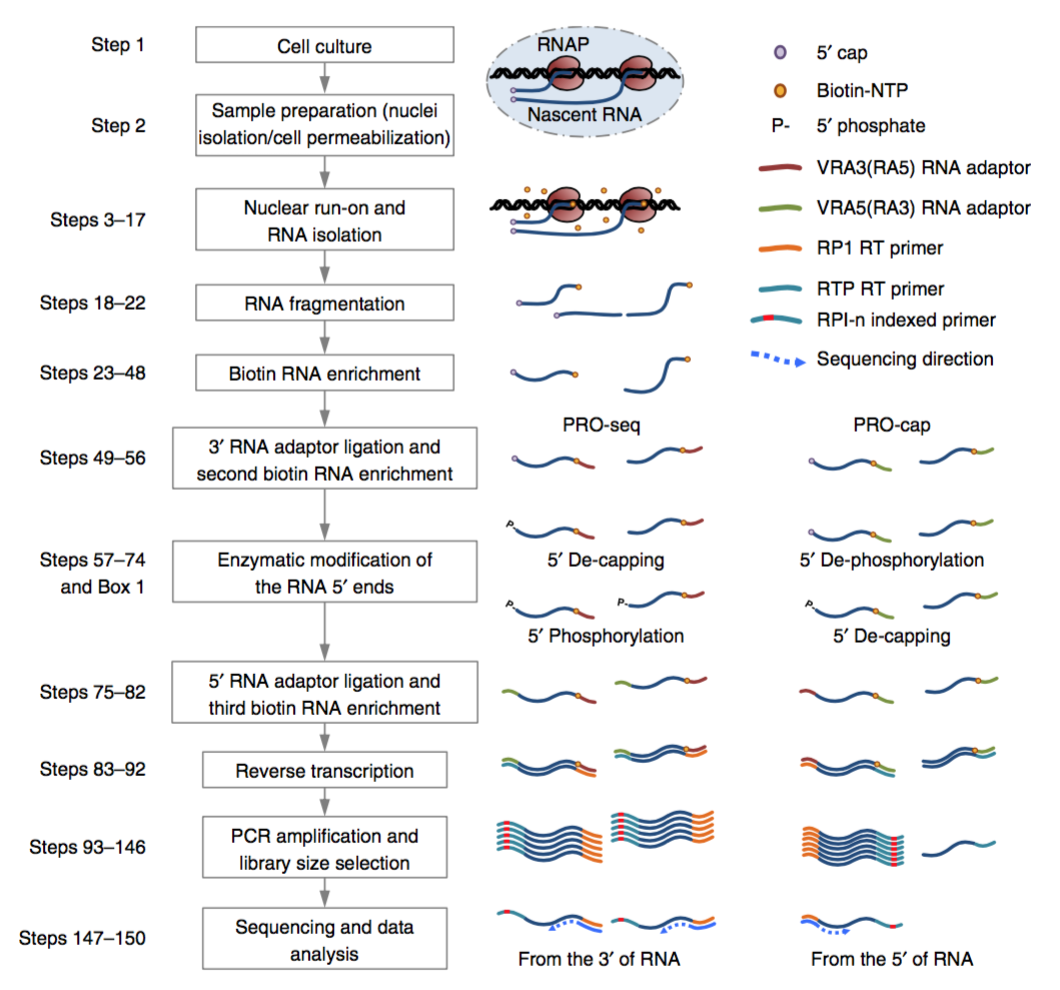
Figure. PRO-seq workflow (Mahat, Kwak et al. 2016)
Reference
Mahat, D. B., H. Kwak, G. T. Booth, I. H. Jonkers, C. G. Danko, R. K. Patel, C. T. Waters, K. Munson, L. J. Core and J. T. Lis (2016). "Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq)." Nat Protoc 11(8): 1455-1476.

Jie Zhang
j dot zhang dot 27 at warwick dot ac dot uk
