Influenza
Described as the 'Last Great Plague' of humankind, both seasonal and pandemic influenza continue to pose major challenges both in terms of optimal response, and understanding disease dynamics. How do new strains emerge and why? When should we shut schools? How effective are antiviral agents? Such questions require advanced methods for modelling and analysis of multiple data sources.
On this page we focus on our work about Pandemic Influenza; we also work on Seasonal Influenza.
Influenza pandemic predictability
Despite influenza pandemics being rare events, with it currently being nearly impossible to predict the next influenza emergence event, it may be the case that the virus itself provides us with outbreak signals that should prompt us to be more prepared. Specifically, knowing whether or not influenza pandemic occurrences are uniformly random in time can inform the intervention strategies that would be best suited to reduce the risk of further pandemic events.
To investigate the predicatability of influenza pandemics, we analysed the time periods between probable influenza pandemics since 1700. We depict below three alternative timelines of reputed historic influenza pandemics that underwent consideration:
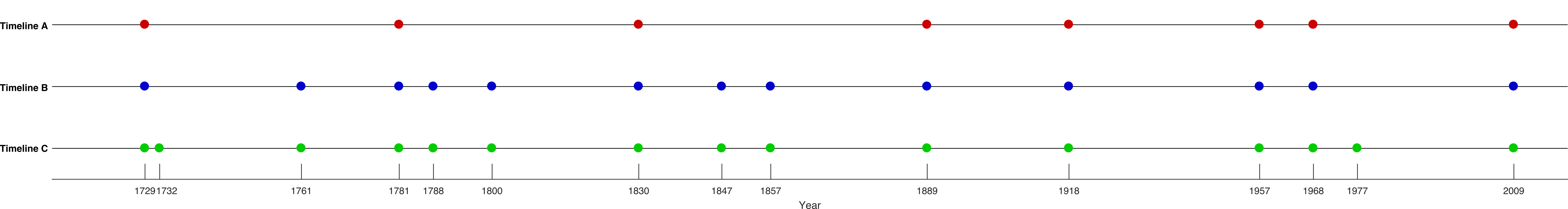
Figure: Graphical timelines for each of the proposed influenza pandemic histories. Dots denote outbreaks considered to be pandemics within the respective timelines.
Using a Bayesian model selection approach we compared plausible model hypotheses regarding the mechanisms driving influenza pandemic occurrences. We found the weight of evidence to favour influenza pandemic emergence being history-dependent, rather than a memoryless process. Conclusions drawn may allow quantitative predictions for number of expected pandemics in a specified time period to be made.
Though the modelling approach taken relies upon limited data, so is uncertain, it provides cheap, safe and direct evidence relating to pandemic emergence, a field where indirect measurements are often made at great risk and cost.
Publications
- Hill EM, Tildesley MJ & House T (2017) How predictable are flu pandemics? Significance 14(6): 28-33. doi: 10.1111/j.1740-9713.2017.01090.x
- Hill EM, Tildesley MJ & House T (2017) Evidence for history-dependence of influenza pandemic emergence. Sci. Rep. 7: 43623. doi: 10.1038/srep43623
H1N1 pandemic
 The world-wide H1N1 pandemic of 2009 was the first influenza pandemic since 1968, and hence the first influenza pandemic since predictive modelling was able to inform policy. Upto 2009, the vast majority of modelling papers and public-health planning was focused towards avian-dervied H5N1 influenza spreading from S.E.Asia. H5N1 has the potential to cause substantial loss of life, with very high mortality levels observed. In reality, the 2009 pandemic began in Mexico, and despite initial concerns, it proved to be rather mild; in fact, although there was little immunity in the population again H1N1, there were fewer fatalities from this pandemic than during a normal 'flu season.
The world-wide H1N1 pandemic of 2009 was the first influenza pandemic since 1968, and hence the first influenza pandemic since predictive modelling was able to inform policy. Upto 2009, the vast majority of modelling papers and public-health planning was focused towards avian-dervied H5N1 influenza spreading from S.E.Asia. H5N1 has the potential to cause substantial loss of life, with very high mortality levels observed. In reality, the 2009 pandemic began in Mexico, and despite initial concerns, it proved to be rather mild; in fact, although there was little immunity in the population again H1N1, there were fewer fatalities from this pandemic than during a normal 'flu season.
 The timing and mildness of the H1N1 pandemic raised a number of issues. In a severe pandemic it is likely that most cases will have symptoms and therefore seek medical advice or in some other way be recorded; however for a mild pandemic it is unclear what proportion of people have sufficiently severe symptoms to be recorded. Or, to put in another way, are there current few cases (most of whom have symptoms) or are there many cases few of whom have symptoms. In addition, the epidemic began in the late spring and persisted through the summer, as this is not the usual 'flu season in the northern hemisphere it was unclear whether a larger epidemic with more severe cases could be expected in the winter.
The timing and mildness of the H1N1 pandemic raised a number of issues. In a severe pandemic it is likely that most cases will have symptoms and therefore seek medical advice or in some other way be recorded; however for a mild pandemic it is unclear what proportion of people have sufficiently severe symptoms to be recorded. Or, to put in another way, are there current few cases (most of whom have symptoms) or are there many cases few of whom have symptoms. In addition, the epidemic began in the late spring and persisted through the summer, as this is not the usual 'flu season in the northern hemisphere it was unclear whether a larger epidemic with more severe cases could be expected in the winter.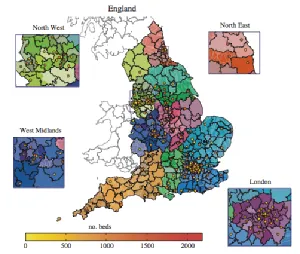
Members of the (then called) Warwick Infectious Disease Epidemiology Research (WIDER) group had worked on pandemic planning issues before, either considering pandemic influenza or bio-terrorist smallpox release. We were therefore happy to analyse the UK and US data as it was being collected and to feed back findings to the relevant government agencies. In particular: Matt Keeling was a member of the UK's Dept of Health SPI-M (Scientific Pandemic Influenza - Modelling) group and provided input to the Dept of Health through several groups; Leon Danon was temporarily based in Harvard (USA) during the pandemic and assisted with their analysis of the US situation; Thomas House analysed the impact of school closures on the pandemic in particular to alleviate the burden on intensive care wards.
Publications
-
Black AJ, House T, Keeling MJ & Ross JV (2013) Epidemiological consequences of household-based antiviral prophylaxis for pandemic influenza. J. Roy. Soc. Interface 10(81): 20121019. doi: 10.1098/rsif.2012.1019
- House T, Inglis N, Ross JV, Wilson F, Suleman S, Edeghere O, Smith G, Olowokure B & Keeling MJ (2012) Estimation of outbreak severity and transmissibility: Influenza A(H1N1)pdm09 in households. BMC Medicine 10: 117. doi: 10.1186/1741-7015-10-117
- House T, Baguelin M, van Hoek AJ, White PJ, Sadique Z, Eames K, Read JM, Hens N, Melegaro A, Edmunds WJ & Keeling MJ (2011) Modelling the impact of local reactive school closures on critical care provision during an influenza pandemic. Proc. Roy. Soc. B 278(1719): 2573-2760. doi: 10.1098/rspb.2010.2688
- Keeling MJ & White PJ (2011) Targeting vaccination against novel infections: risk, age and spatial structure for pandemic influenza in Great Britain. J. Roy. Soc. Interface 8(58): 661-670. doi: 10.1098/rsif.2010.0474
- Miller JC, Danon L, O'Hagan JJ, Goldstein E, Lajous M & Lipsitch M (2010) Student Behavior during a School Closure Caused by Pandemic Influenza A/H1N1. PLoS ONE 5(5): e10425. doi: 10.1371/journal.pone.0010425.
- Goldstein E, Cowling BJ, O'Hagan JJ, Danon L, Fang VJ, Hagy A, Miller JC, Reshef D, Robins J, Biedrzycki P & Lipsitch M (2010) Oseltamivir for treatment and prevention of pandemic influenza A/H1N1 virus infection in households, Milwaukee, 2009. BMC Infect. Dis. 10: 211. doi: 10.1186/1471-2334-10-211
- Sigmundsdottir G, Gudnason T, Olafsson O, Baldvinsdottir GE, Atladottir A, Love A, Danon L & Briem H (2010) Surveillance of influenza in Iceland during the 2009 pandemic. Eurosurveillance 15(49): pii=19742. doi: 10.2807/ese.15.49.19742-en
- Lajous M, Danon L, Lopez-Ridaura R, Astley CM, Miller JC, Dowell SF, O'Hagan JJ, Goldstein E & Lipsitch M (2010) Mobile Messaging as Surveillance Tool during Pandemic (H1N1) 2009, Mexico. Emerg. Inf. Dis. 16(9): 1488-1489. doi: 10.3201/eid1609.100671
- Danon L, House T & Keeling MJ (2009) The role of routine versus random movements on the spread of disease in Great Britain. Epidemics 1(4): 250-258. doi: 10.1016/j.epidem.2009.11.002
- Keeling MJ & Danon L (2009) Mathematical modelling of infectious diseases. British Med. Bull. 92: 33-42. doi: 10.1093/bmb/ldp038
- House T & Keeling MJ (2009) Household structure and infectious disease transmission. Epidemiol. Infect. 137(5): 654-661. doi: 10.1017/S0950268808001416
- Lipsitch M, Lajous M, O'Hagan JJ, Cohen T, Miller JC, Goldstein E, Danon L, Wallinga J, Riley S, Dowell SF, Reed C & McCarron M (2009) Use of Cumulative Incidence of Novel Influenza A/H1N1 in Foreign Travelers to Estimate Lower Bounds on Cumulative Incidence in Mexico. PLoS ONE 4(9): e6895. doi: 10.1371/journal.pone.0006895
- Wearing HJ, Rohani P & Keeling MJ (2005) Appropriate models for the management of infectious diseases. PLoS Med. 2(7): e174 doi: 10.1371/journal.pmed.0020320
Funded by: MRC, EPSRC
SBIDER people involved:
Internal collaborators:
External collaborators:
Thomas House (Manchester)
Jon Edmunds (LSHTM)
Josh Ross (Adelaide)
Jon Read (Lancaster)
Ken Eames (formerly LSHTM)
