Research
In vitro single-molecule studies of virus replication
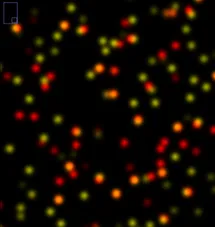
During an infection, a virus enters a cell, disassembles into functional components, and harnesses the cellular machinery to produce multiple copies of itself; these new copies then spread to neighboring cells. Much of our knowledge of this process relies on data obtained using ensemble methods that report on the averaged properties of millions of molecules. However, using single-molecule techniques, we can now measure the structural and functional properties of single replication complexes in real-time, revealing unique details that may be difficult to see using traditional biology or biochemistry methods.
We combine biochemistry and virology with single-molecule assays like Förster resonance energy transfer (FRET) to investigate the mechanisms of replication (RNA binding, initiation, elongation and termination) of RNA viruses.
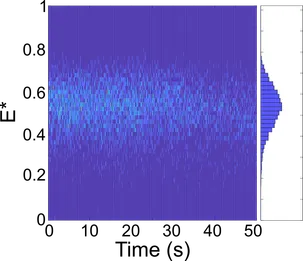
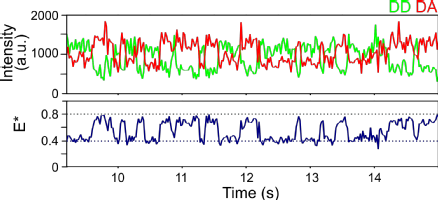
In vivo studies of virus replication
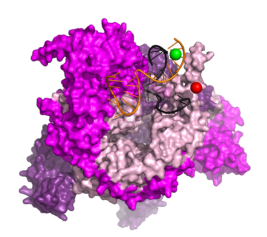
We study the morphology and cellular interactions of influenza and coronaviruses using super-resolution fluorescence microscopy techniques such as dSTORM, PALM and single-molecule FISH. Understanding the interactions between viruses and the infected cell is vital to combating viral disease, and allows us to directly assess the actions of antiviral inhibitors in vivo.
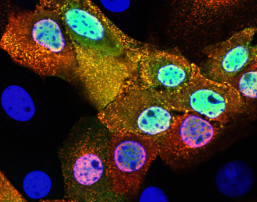


Virus detection
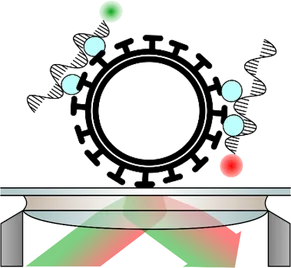
Infectious diseases caused by RNA viruses such as influenza, Ebola, SARS-CoV-2 and hepatitis C represent a huge global public health concern. The rapid and accurate identification of viral pathogens is needed to help prevent transmission, set up appropriate therapy, monitor response to treatment and lead to efficient disease management and control.
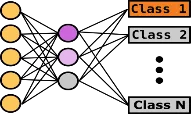
We develop rapid and sensitive fluorescence-based diagnostic assays to identify RNA viruses in complex biological samples, combined with machine learning algorithms for data analysis. These assays have the potential for medical impact as the advantages over traditional diagnostic tests include specificity, sensitivity, speed and ease-of-use.