All Data
Tue 22 March 2005 - first checks using large cell
A six sample experiment was devised. The experiment was performed with repetition to give a total of twelve samples. Preparation and measurement of the linear dichroism spectra were done in a pre-devised order to facilitate tracking of the experiment and to ensure statistical validity by reducing systematic error and/or systematic contamination.
20 mg of soybean lipid was placed into a vial with 4 mL of 5 mM phosphate buffer. The soybean lipid was dissolved by vortexing (Rotamixer Deluxe). This solution was extruded through a polycarbonate membrane (19 mm diameter with pores of diameter 100 nm) eleven times and placed into a new vial.
1 mg of gramicidin was placed into a new vial and dissolved with a few drips of methanol. Both the soybean lipid solution and the gramicidin solution were placed into the refrigerator for 30 minutes. 1 mL of the soybean lipid solution was added tiny drop by tiny drop into the gramicidin-methanol vial so that the gramicidin did not precipitate out of solution.
The experiment
Types of samples tested
- W de-ionized water
- B phosphate buffer
- G gramicidin-methanol solution
- OL soybean lipid solution without extrusion (original liposomes)
- EL extruded soybean lipid solution (extruded lipsomes)
- M extruded mixture of soybean lipid and gramicidin
Design of experiment
| Label | Sample | Buffer/mL | Lipid/mg | Gramicidin/mg |
|---|---|---|---|---|
| 1 | W | 4 | 0 | 0 |
| 2 | B | 4 | 0 | 0 |
| 3 | M | 4 | 20 | 1.0119 |
| 4 | OL | 4 | 20 | 0 |
| 5 | G | 4 | 0 | 1.1842 |
| 6 | EL | 4 | 20 | 0 |
| 7 | W | 4 | 0 | 0 |
| 8 | B | 4 | 0 | 0 |
| 9 | G | 4 | 0 | 1.2087 |
| 10 | OL | 4 | 20 | 0 |
| 11 | M | 4 | 20 | 1.0072 |
| 12 | EL | 4 | 20 | 0 |
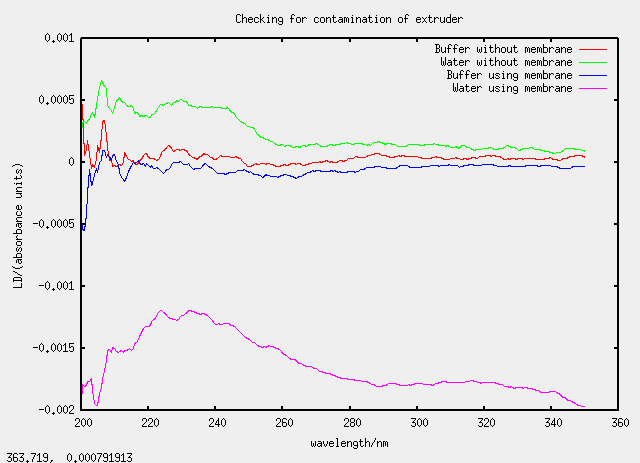
Thu 23 March - checking contamination of extruder
The experiment
Design of experiment
| Label | Lipid/mg | Liquid/mL | Membrane? |
|---|---|---|---|
| alpha | 20 | 4 (buffer) | no |
| beta | 20 | 4 (water) | no |
| gamma | 20 | 4 (buffer) | yes |
| delta | 20 | 4 (water) | yes |
Something's gone wrong with the fourth sample
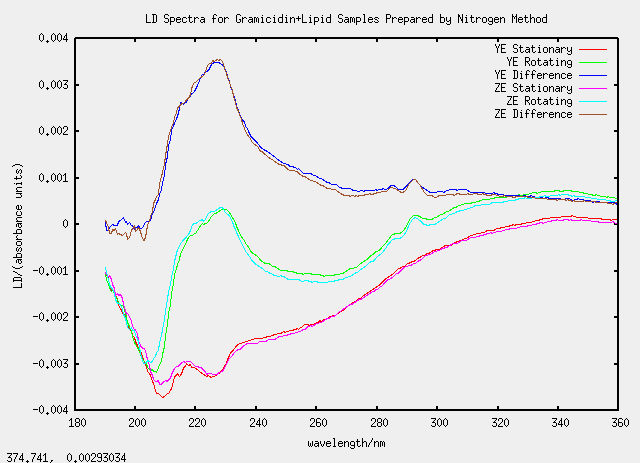
Thu 31 March - nitrogen method
2.5 mg of soybean lipid was placed into a vial and 0.5 mg of gramicidin was added. 2 mL chloroform was added to create a homogeneous mixture. The chloroform was evaporated with a stream of air over night. The film left at the bottom of the vial was resuspended in 1 mL of phosphate buffer by vortexing. The mixture was extruded through a polycarbonate membrane of pore diameter 100 nm a total of 11 times. The extruded mixture was subjected to five cycles of freeze and thaw, using liquid nitrogen for rapid freezing, before being left to refrigerate over night.
| Label | Lipid/mg | Gramicidin/mg |
|---|---|---|
| X | 2.5 | 0 |
| Y | 2.6 | 0.5704 |
| Z | 2.6 | 0.5538 |
Samples YE and ZE were nice as they agreed well despite being prepared separately.
Tue 12 April - TatABC calibration test
| Label | Lipid/mg | Insert/mg |
|---|---|---|
| buffer | 0 | 0 |
| G1 | 0 | 0.2690 (gramicidin) |
| L1 | 2.5 | 0 |
| T1 | 2.5 | 0.277 (TatABC) |
| G2 | 1.6 | 0.3712 (gramicidin) |
| L2 | 2.5 | 0 |
| G3 | 0 | 0.2233 (gramicidin) |
| T2 | 2.5 | 0.277 (TatABC) |
Thu 21 April - attempt at better mixing
In attempt to give the gramicidin and TatABC a better chance at insertion and to give a more homogeneous distribution of liposomes, some of the samples were evaporated to a film and resuspended in phosphate buffer by brief sonication.
| Label | Chloroform added? |
|---|---|
| CG2 | yes, to G2 |
| NL1 | no, L1 evaporated and resuspended |
| NG2 | no, G2 evaporated and resuspended |
| NT1 | no, T1 evaporated and resuspended |
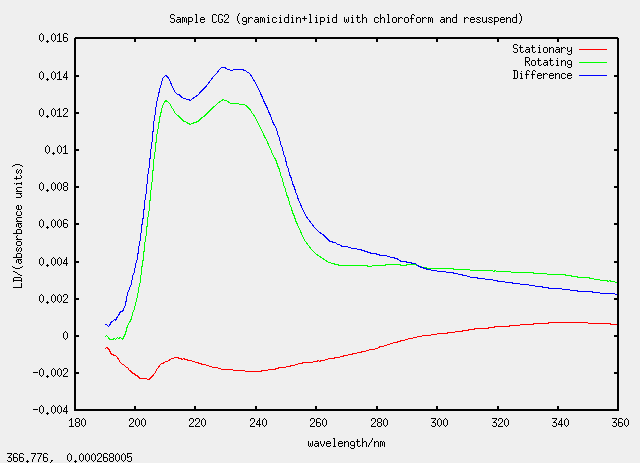
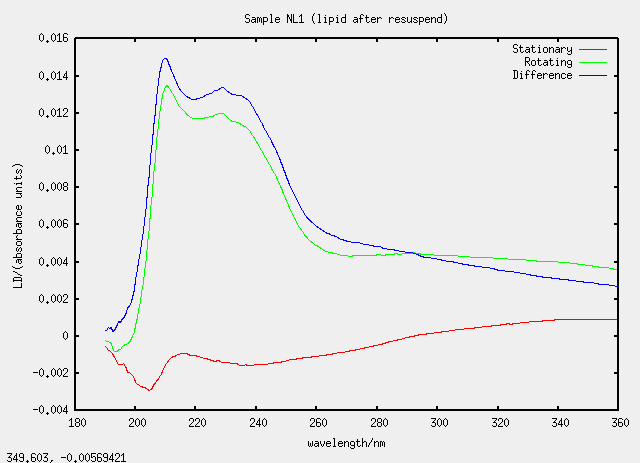
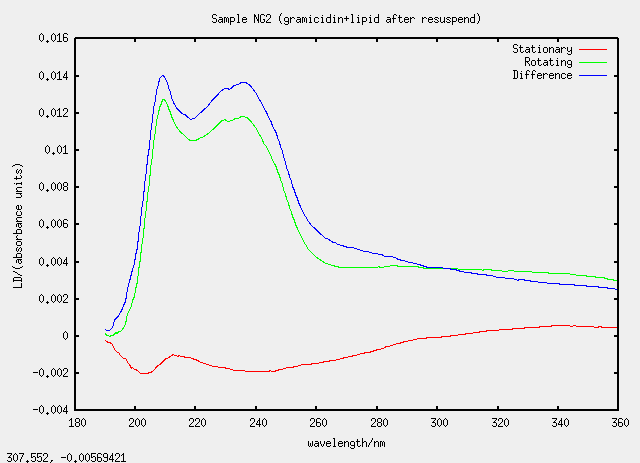
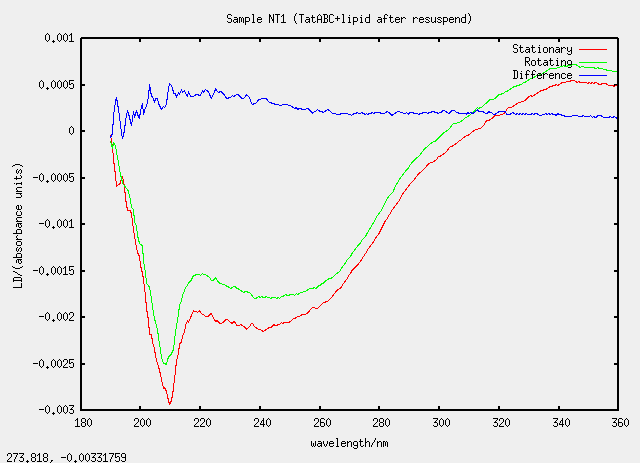
Something silly and strange is going on
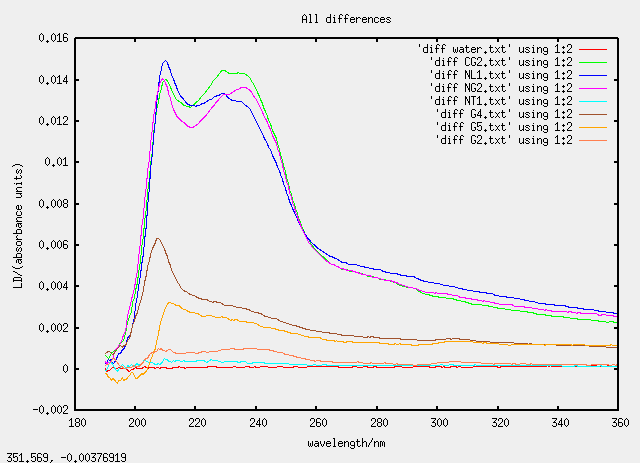
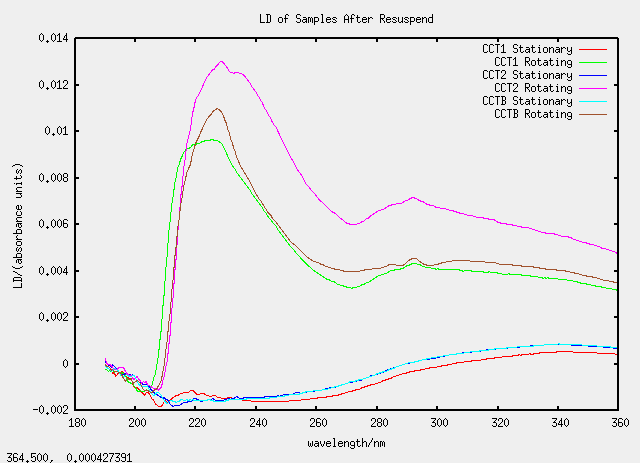
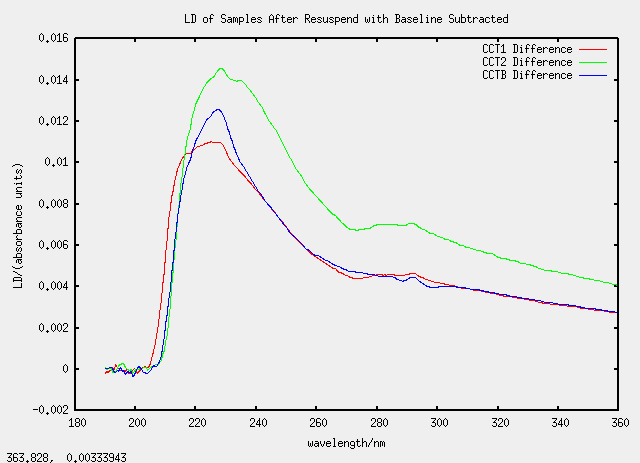
Sat 23 April - chloroform dissolving
All the CC samples (C for chloroform and duplicated for emphasis) are all based on TatABC samples.
| Label | Description | (Chloroform added)/micro-litre |
|---|---|---|
| CCT1 | 100 micro-litres of T1 | 1400 |
| CCT2 | 150 micro-litres of T2 | 1400 |
| CCTB | 150 micro-litres of T1b | 1400 |
| CCQT | special (see below) | n/a |
| CCRU | special | n/a> |
A white substance was observed to form on the surface of the mixture on the addition of more than 1 mL of chloroform so samples CCQT and CCRU were prepared. CCQT (QT standing for "Quick Test") was prepared with 150 micro-litres of T2 and 1400 micro-litres of chloroform. The translucent liquid on the bottom was removed by pipette to form sample CCRU (the letters QT have been advanced once in the alphabet). CCQT was topped up with 50 micro-litres of buffer. All dispersions in chloroform were sonicated briefly to encourage mixing.
As we are getting used to now, the graphs are strange