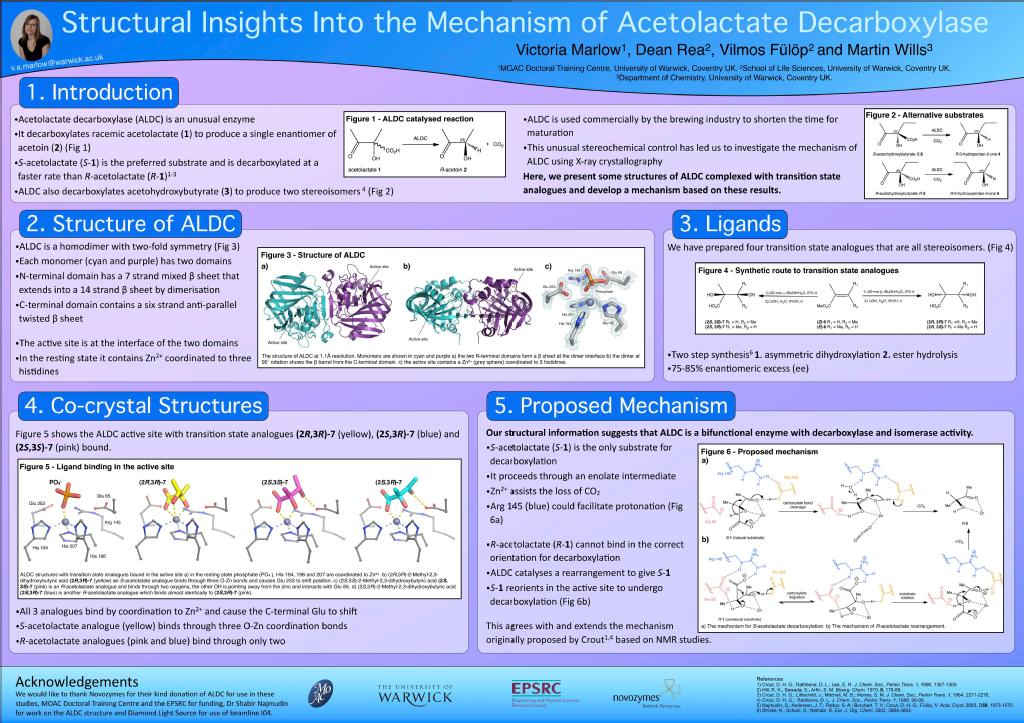
Structure and Mechanistic Studies of Acetolactate Decarboxylase
Acetolactate Decarboxylase (ALDC) is a bacterial enzyme involved the butanediol biosynthetic pathway. It catalyses the conversion of both enantiomers of acetolactate into enantiomerically pure R-acetoin as shown in scheme 1.
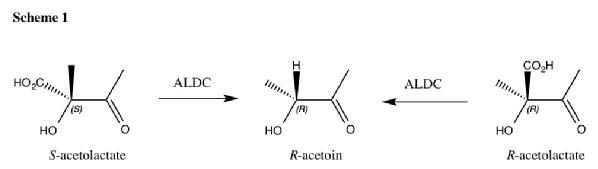
It is unusual that the enzyme is able to use both enantiomers of substrate yet produce only one enantiomer of product. The molecular mechanism of catalysis is unknown and is the basis of my PhD.
The structure of Acetolactate decarboxylase from Bacillus brevis has been solved using X-ray crystallography by the Fulop group. The structure shown in Figure 1 has been deposited in the PDB (accession code 4BT2).
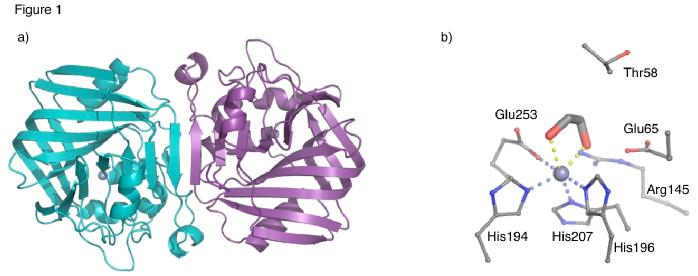
ALDC is a mixed α/β protein that forms a homo-dimer with two-fold symmetry. Each monomer contains two domains, the N-terminal domain contains a 7 strand mixed β-sheet, which extends to form a 14 strand β-sheet upon dimerisation. The C-terminal domain contains a β-barrel structure that is formed from a six strand antiparallel β-sheet that folds over to form the barrel structure. The active site is located at the interface between the two domains. In the B. brevis structure, shown in Figure 1b, the Zn2+ ion is coordinated to three highly conserved histidines (194,196 and 207) and a glutamate (Glu253) from the C-terminal tail. The structure also shows ethylene glycol (EG) bound to metal ion. EG was present in the crystallisation conditions and is a product analogue. Also in close proximity are Thr58, Arg145 and Glu65, which are also highly conserved and possibly play a role in catalysis or stabilisation of intermediates.
During my PhD I sought to gain insight into the molecular mechanism of the ALDC catalysed reaction by using a combination of kinetic and structural techniques. Bacillus subtilis alsD encoding ALDC was cloned into an expression vector and a series of active site mutants were prepared. The activity of mutant AlsD were determined using a circular dichroism based assay, which identified that the two active site glutamates and a basic residue are required for catalysis. A series of chiral transition state analogues were prepared in a two-step synthesis to give enantiomerically enriched 2,3-dihydroxy-2-methylbutanoic acid in reasonable yields. Three of the compounds were identified as competitive inhibitors, co-crystallised with Bacillus brevis ALDC and structures solved to 1.1-1.6 Å. These structures, coupled with inhibition studies and site-directed mutagenesis, provide an improved understanding of the molecular processes involved in the stereoselective decarboxylation of acetolactate. Further details on this work can be found in the following ACS Chemical Biology publication and poster that was presented at the CCP4 Northern Protein Structures conference 2012.
'Structure and Mechanism of Acetolactate Decarboxylase', Victoria A. Marlow, Dean Rea, Shabir Najmudin, Martin Wills, and Vilmos Fülöp, ACS Chemical Biology, 2013, 8, pp tba. DOI: 10.1021/cb400429h. Publication Date (Web): August 14, 2013 (Articles).
The ALDC structures that I have solved during my PhD have been deposited in the PDB under accession codes 4BT3, 4BT4, 4BT5, 4BT6 and 4BT7.