Alvin Teo
Welcome to my page! My name is Alvin and I am from Malaysia. I am in the Roper Group at the C10 Structural Biology Lab, School of Life Sciences, but currently undertaking my academic secondment at Aston University, under the supervision of Prof. Corinne Spickett and Prof. Andrew Pitt. My main research interest centres on the study of various membrane proteins involved in cell division and peptidoglycan biosynthesis in bacteria, focusing on the Gram negative model organism - E. coli.
Research Sypnosis
About one third of the gene transcripts in the natural world encodes for proteins that are embedded within a lipid membrane, and they represent the target for over 50% of marketed drugs. However, this repertoire of membrane proteins are relatively underexplored due to the difficulties in extraction and solubilisation in their active forms for functional and structural studies. Traditional approaches using detergent for the membrane proteins solubilisation can be denaturing due to the sequestration of membrane phospholipids surrounding the proteins. Furthermore, the membrane proteins solubilised in detergent micelles are not presented in their native lipidic environment for subsequent biophysical and biochemical analyses. In my project, I explore the application of a novel detergent-free approach to encapsulate the membrane proteins together with their annular lipidic components directly from native membrane environment (in a pH-dependent manner) into nanoscale lipid discs stabilised by a copolymer of styrene maleic acid (SMA).
My research looks into aspect of bacterial cell wall biosynthesis and its inherent connection to cell division. Before a bacterial cell is ready to divide, new cell wall has to be formed. This is a complex yet well-coordinated process, with a plethora of proteins interacting in a cooperative and transient manner on both sides of the cell membrane. Peptidoglycan, the major structural component of the bacterial cell wall is a polymeric mesh consisting of alternating units of N-acetylmuramic acid and N-acetylglucosamine, being cross-linked via stem peptide bridges. The emphasis is placed on the membrane-associated stage for the biosynthesis of peptidoglycan and my project addresses the challenges of integral membrane protein purification and their extraction and solubilisation in active form with native lipids in the total absence of detergents. Being the Achilles’ heel of a bacterial cell, the cell wall has been the major target for the discovery of antibiotics over many decades. As part of the global effort to tackle the escalating antimicrobial resistance issue, a range of bioanalytical studies facilitated by this SMA lipid particle (SMALP) system may provide the vital information on the structure and function of many of these membrane proteins to underpin the development of next generation antimicrobial drugs.
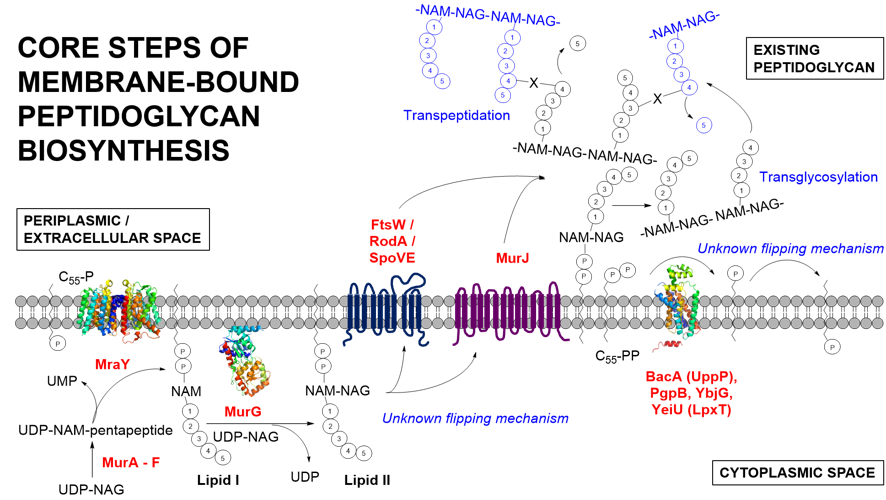
The SMALP approach has been recently successfully applied to a number of proteins involved in bacterial cell division and peptidoglycan biosynthesis, including an integrated membrane type II phosphatidic acid phosphatase of Escherichia coli – PgpB. PgpB is demonstrated to participate in the metabolism of undecaprenyl phosphate (C55-P) with several other phosphatases like BacA/UppP and YbjG. The C55 lipid carrier serves as the essential shuttle molecule for the biosynthesis of many cell wall polysaccharides (including peptidoglycan). The purified protein in SMALP is properly folded as demonstrated by circular dichroism analysis with the characteristic α-helical negative minima at 208 and 222 nm. Current efforts are directed towards developing a functional biochemical assay to assess the activity of this critical enzyme.
In addition, methodology development for the profiling of lipidic components co-extracted with PgpB and several other cell division membrane proteins via liquid chromatography-mass spectrometry (LC-MS) is in active progress for the study of specific protein-lipid interaction.
Academic History
2013 (Sep) - Present:
- PhD (on-going), University of Warwick
'Methodologies for cell division membrane protein production, purification and characterisation'
Supervisor: Dr. David Roper
- Modules taken: CH908 Mass Spectrometry, CH911 Chromatography and Separation Science, CH926 Molecular Modelling
- Attended EMBO practical course - Protein Expression, Purification, and Characterisation (PEPC9), Hamburg, 8-16 September 2014 (by selection)
- Awarded best poster prize in the 2nd Midlands Molecular Microbiology Meeting, University of Nottingham, 14-15 October 2015
- Publication: Teo and Roper (2015) Core steps of membrane-bound peptidoglycan biosynthesis: recent advances, insight and opportunities, Antibiotics: 4, 495-520
2006 - 2008:
- BSc (Hons), Biotechnology, Northumbria University
'The cloning of amidase gene (>gi|39934541) from Rhodopseudomonas palustris CGA009 into pCR®-Blunt vector with subsequent transformation of recombinant Escherichia coli cells'
Supervisors: Prof. Gary Black, Dr. Simon Charnock (Prozomics Ltd.)
Working History
2011 (Nov) - 2013 (Jul):
- Wipro Skin Research & Innovation Centre, Research & Development Department, Malaysia
Product Development Chemist - Commercialised products include: Aiken Antibacterial Shower Crème, Dashing Face & Body Wash, Eversoft Beauty Body Wash, Gervene Family & Luxury Shower Creme, Safi Toothpaste & Mouthwash
2010 (May) - 2011 (Oct):
- Synthomer - Technical Service Department, Malaysia
Technologist
2008 (Nov) - 2010 (Sep):
Email: A.C.K.Teo@warwick.ac.uk
Address:
MOAC DTC
Senate House
University of Warwick
Coventry
CV4 7AL
School of Life Sciences
Gibbet Hill Campus
University of Warwick
Coventry
CV4 7AL

