SOMspec (formerly SSNN, Secondary Structure Neural Network)
SOMspec (Self-Organising Map for spectroscopy), formerly called SSNN, (secondary structure neural network) is a self organising map software used to cluster data from circular dichroism or infrared spectra to estimate protein secondary structures developed by Vincent Hall, Anthony Nash, Alison Rodger (now at Macquarie) and Nikola Chmel.
The performance of SOMspec is at least as good as other available protein CD structure-fitting algorithms and it shows superior performance when the exact protein concentration is unknown. SOMspec can also be used to predict protein secondary structure form infrared and Raman data.
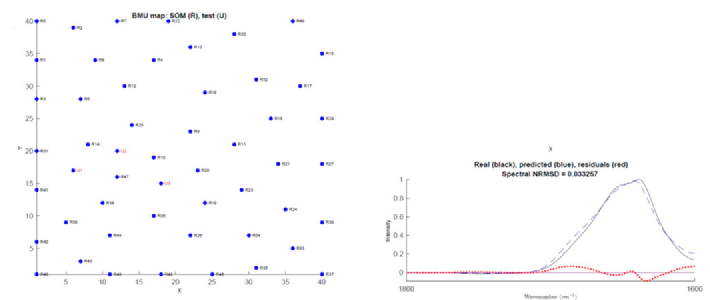
Image: (left) Phosphoglycerate kinase SOMSpec output from 50-protein FTIR film reference set for a relatively poor quality example. (right) SOMspec user interface.
SOMspec work has been published in the following articles:
Elucidating protein secondary structure with circular dichroism and a neural network - Journal of Computational Chemistry
SSNN, a method for neural network protein secondary structure fitting using circular dichroism data - Analytical Methods
Protein Secondary Structure Prediction from Circular Dichroism Spectra Using a Self‐Organizing Map with Concentration Correction - Chirality
Circular dichroism for secondary structure determination of proteins with unfolded domains using a self-organising map algorithm SOMSpec - RSC Advances
Biophysical characterization of a protein for structure comparison: methods for identifying insulin structural changes - Analytical Methods
SOMSpec as a General Purpose Validated Self-Organising Map Tool for Rapid Protein Secondary Structure Prediction From Infrared Absorbance Data - Frontiers
Licence:
A free download of the software is available for academic use only. Please contact Nikola Chmel for details.
If you are interested in using the Software commercially, please contact Warwick Ventures Limited ("WVL"), the technology transfer company of the University, to negotiate a licence.
