Theorectical Project
Molecular Dynamics Investigation of DNA-Protein Interactions Involved in Transcriptional Regulation.
Dr Rebecca Notman, Center for Scientific Computing.
Theorectical & Computational Chemistry - Rebecca Notman Group
The resulting poster outlining the work undertaken in the project can be found here![]() .
.
A presentation given at the 2010 Annual MOAC conference outlining the findings of the project can be found here![]() .
.
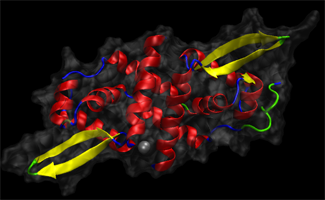
Project Background:
Specific interactions between proteins and DNA are fundamental to many biological processes including
transcriptional regulation. It is therefore important to elucidate the mechanisms that control protein-DNA recognition
in order to understand these processes and identify ways to regulate them for biomedical applications.
SmtB is a DNA-binding protein that is involved in zinc homeostasis. In the absence of zinc the protein binds to
DNA, where it represses the transcription of SmtA, a protein that binds excess zinc. When zinc binds to SmtB,
transcription of the gene for SmtA is switched on and the zinc level in the cell is reduced. Recently, ionmobility
mass spectrometry experiments have shown that zinc-bound and unbound SmtB is present in two main
conformations (extended and compact); however in the bound state, the equilibrium shifts towards the more
compact conformation. While it is clear that zinc binding induces a conformational change in SmtB, it is not known if
the zinc-bound SmtB dissociates from DNA or remains bound to it in a way that allows transcription to occur.
Molecular dynamics (MD) simulation is an approach that shows great promise for the study of DNA-protein
interactions.
This miniproject will use MD simulations to explore the structure of SmtB and its interactions with
DNA. Simulations will be carried out at the atomistic level using the CHARMM forcefield as implemented within
Gromacs. The aim is to gain insights into the effects of zinc on the structure of SmtB, and the relationship
between protein structure and DNA-binding properties. The simulations will complement the experimental approach
and will be validated against the existing experimental data.
Further Reading:
MacKerell, A. D. & Nilsson, L. (2008). Molecular Dynamics simulations of nucleic acid-protein complexes. Curr. Opin. Struct. Biol. 18, 194-199.
VanZile, M. L., Chen, X. H. & Giedroc, D.P. (2002). Allosteric negative regulation of smt O\P binding of the zinc sensor, SmtB, by metal ions: A coupled equilibrium analysis. Biochemistry. 41, 9776-9786.
(*Project Background adapted from the original MOAC mini-project proposal)

