OD Walkthrough

Version: 1.0a
Contacts: gareth.price@warwick.ac.uk; a.marsh@warwick.ac.uk
Example 1: Statin + HMG-CoA Reductase
You can follow along with this example by downloading the initial PDB from here. Before starting, watch the video tutorial provided by the authors of AutoDock Vina, here.
Preparing the files
We first need to split the crystal structure of the complex into separate receptor and ligand files. Open up the file 1T02.pdb in your favourite text editor and look at the file. The first section are REMARKS about the structure:
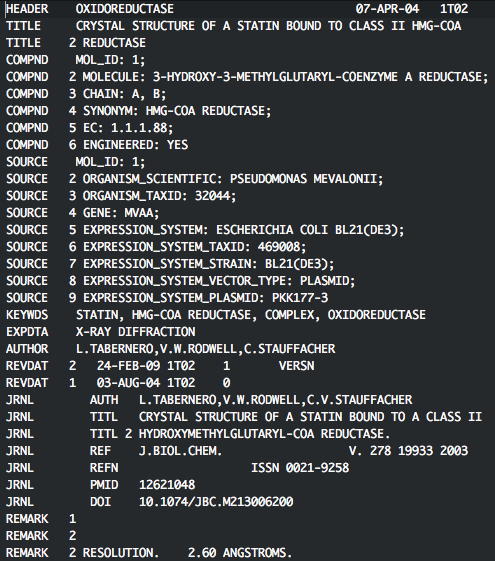
Delete all the header lines, up to line 765 which is the first atom of the structure.
Copy all lines with ATOM at the start into its own file, named hmg_coa.pdb. Include the TER statement in line 5529. Then, copy the subsequent lines starting with HETATM and LVA in the the fourth column into its own file, LVA.pdb:
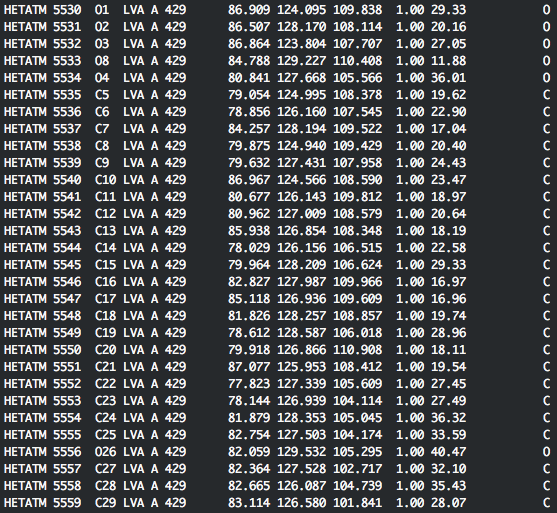
Now, put hmg_coa.pdb into a 'receptor' folder and LVA.pdb into a 'pdb' folder. Next, we need to prepare the receptor protein (hmg_coa.pdb).
Open up AutoDockTools, and follow the steps in the image below. Make sure to remember the coordinates and dimensions. Make sure Spacing (Angstrom) is set to 1.000.
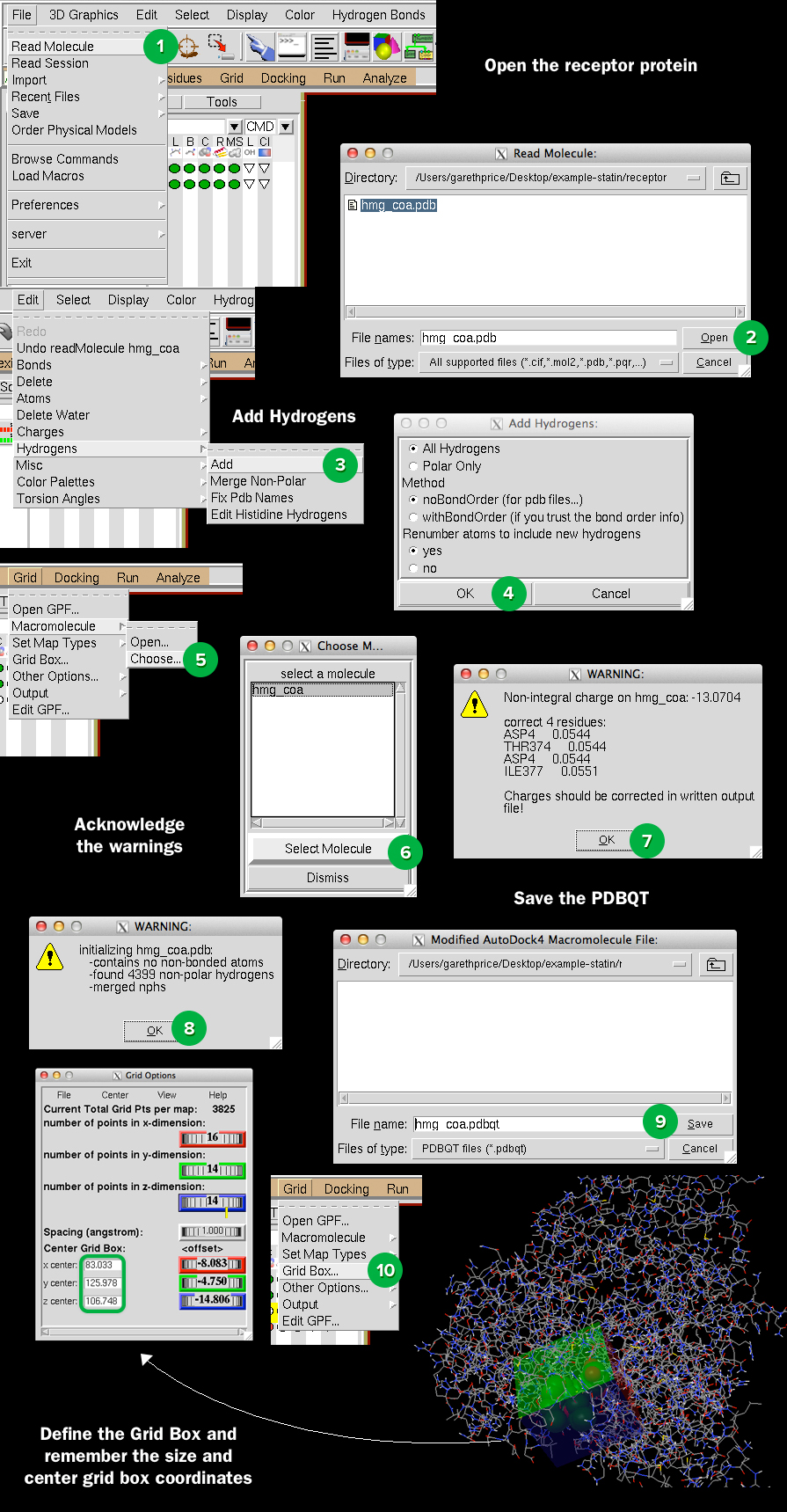
Open a new file and enter the box coordinates and dimensions such as:
center_x = 83.033
center_y = 125.978
center_z = 106.748
size_x = 16
size_y = 14
size_z = 14
Save it as hmg_coa.conf.
You should have a folder that looks like:
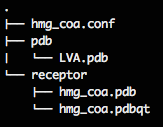
Running ODScreen
We can now run ODScreen. First, let's check that everything is installed properly. In terminal, cd to the Protocol Folder (i.e. the folder with odscreen.py, odparam.py etc.) and run python odcheck.py. Check that there are no Failures, and follow the installation guides if there are. Now, we can run the screening protocol. Navigate to the folder where hmg_coa.conf is located (from before). Now we can run odscreen.py. Make sure you use the correct path to the odscreen.py file.
python /protocolfolder/odscreen.py -d . -r hmg_coa -i pdb -c hmg_coa.conf
This is the result:
# ----------------------------------------- #
# OPEN DISCOVERY #
# Screening Module #
# ----------------------------------------- #
# Version: 1.0 #
# URL: www.opendiscovery.org.uk #
# Contacts: gareth.price@warwick.ac.uk #
# a.marsh@warwick.ac.uk #
# ----------------------------------------- #
# LigDir: /Desktop/example-statin #
# Receptor Name: hmg_coa #
# Input Type: pdb #
# Conf: hmg_coa.conf #
# Exhaustivness: 20 #
# ----------------------------------------- #
# Time Started: Mon, 22 Jul 2013 00:15:20 #
# ----------------------------------------- #
PDB -> SMILES
Writing smiles/LVA.txt
IMAGES
Writing images/LVA.svg
MOL
Writing mol/LVA.mol
MOL2
Writing mol2/LVA.mol2
MINIMISATION
Minimising LVA
PDBQT PREPARATION
Writing LVA.pdbqt
SCREENING
Processing LVA
EXTRACTING
Processing results/LVA/
PDB -> MOL2
Writing results-mol2/LVA.mol2
SUMMARISING
Summarising LVA
MAKING COMPLEXES
Writing LVA
# ----------------------------------------- #
# FINSHED #
# Time Taken: 231.40 seconds #
# ----------------------------------------- #
The resulting files are:
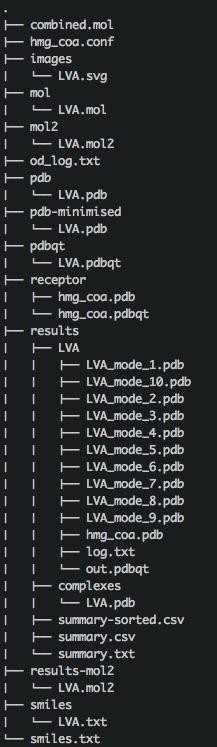
From this point on you can analyse the results. A starting point is to look at the binding energies, sorted and summarised in the sum
