Empirical Validation
ThermoPhyl was validated in practice by pyrosequencing of amplicons designed for optimum specificity and sensitivity for the methanogenic archaeaon Methanosaeta.
PCR primers designed with ThermoPhyl were optimized empirically with gDNA from pure cultures of target and non-target strains and environmental samples. Amplicons (420 bp) from PCR of two different estuarine sediment samples were pooled from three technical and biological replicates after normalization based on picogreen quantification. The key results from pyrosequencing of these amplicons are summarized below.
| Top BLASTP hit |
Site1 |
Site10 |
| Targets |
||
| Methanosaeta_concilii_AF313802 |
2563 |
1674 |
| Methanosaeta_harundinacea_AY970348 |
1825 |
13 |
| Methanosaeta_concilii_VeAc9_AF313803 |
115 |
148 |
| Methanosaeta_thermophila_PT_gb|ABK14360.1 |
9 |
10 |
| Non-targets |
||
| Methanothermobacter_marburgensis_X07794 |
2 |
|
| Methanothermus_fervidus_J03375 |
1 |
|
| Methanococcus_jannaschii_mrtA_U67465 |
1 |
|
| Methanococcoides_burtonii_U22234 |
1 |
|
| Total |
4517 |
1745 |
Table 1. Blastp summary showing specificity of 99.9% (5 non-target sequences out of 6262 total). Sequences derived from pyrosequencing were analyzed by blastp run locally on a custom database containing 44 representative mcrA sequences from pure cultures, including all Methanosaeta strains. The only changes to default parameters were the use of soft-masking (-F “m S”) to enable filtering for low-complexity subsequences during the word seeding phase but not the extension phase of the blastp algorithm.
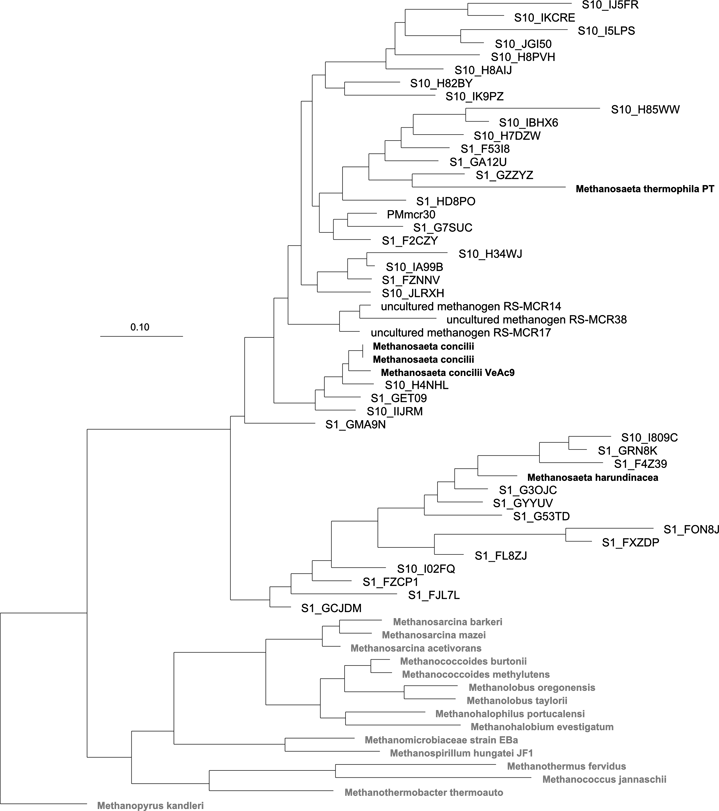
Figure 1. Assay sensitivity illustrated by novel mcrA sequence diversity affiliated with Methanosaeta recovered by ThermoPhyl-generated primers. The tree is a maximum-likelihood phylogenetic reconstruction based on alignment of nucleotides restricted to the amplicon region. Sequences from this study are represented by ‘S1’ or ‘S10’ representing the sampling site and those shown are representatives of the 27 OTUs defined.
