Examples
High-throughput MICs
Manual and robotic 12 point MICs were carried out on 51 novel compounds in DMSO along with 5 known antibiotics (Amikacin, Ticarcillin, Minocyclin , Ri-
fampin, Clarithromycin). In triplicate analysis, 77% of evaluations were within one dilution of the manual read. 23 of the compounds within the library were classied as hits (MIC of <32 mg/ml), robotMIC was able to correctly determine a hit 85% of the time. This is comparable to results achievable from manual screening.
Checkerboards
A Checkerboard between clavulanic acid and amoxicillin was prepared using the liquid handling robot. Figure 2 shows the synergistic interaction between clavulanic acid and amoxicillin against b-lactamase expressing E. coli, with amoxicillin able to inhibit growth once b-lactamases are inhibited.
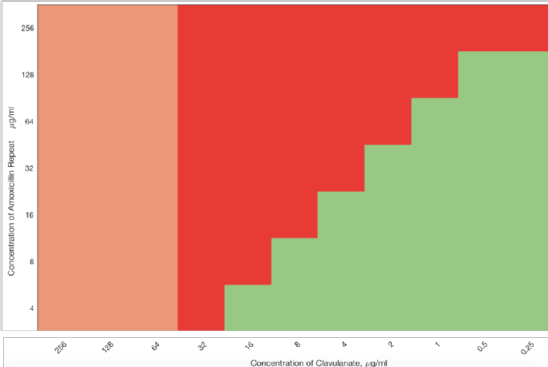 |
| An 7x11 grid checkerboard between clavulanic acid (MIC of 64 mg/ml) and amoxicillin (MIC of >256 mg/ml) in the ESBL-expressing E.coli strain NCTC 13353. Pink indicates inhibition of growth due to the clavulanic acid, green indicates growth and red indicates inhibition of growth due to the amoxicillin in the presence of clavulanic acid. The amount of red illustrates that far more inhibition is possible once the drugs are combined. |
Checkerboards allow for easy identication of synergy between two compounds and can be used to investigate mechanism of action. However due to the number of conditions tested, these assays are slow and labour intensive to carry out by hand. By automating the liquid handling, larger screens can be carried out, allowing greater exploration of the many possible combinations. By reducing the size from 7 x 11 to 4 x 4 (gure 3) , throughput can be increased further.
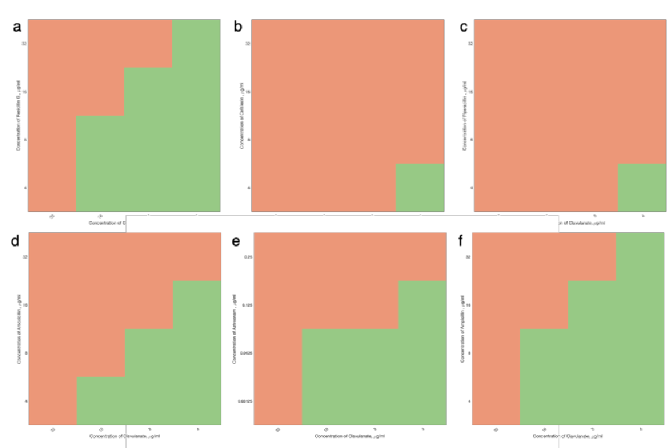 |
| A 4x4 checkers allow increased throughput of synergy studies, typically a very slow process. 6 b-lactams: penicillin G, cephalexin, piperacillin, amoxicillin, aztreonam and ampicillin (a-f respectively ) were tested in synergy with clavulanic acid against ESBL-expressing E.coli strain NCTC 13353. These results are triplicates, which were shown to be highly reproducible. |
Follow-up Work Available
All hits identied by RobotMIC can be rapidly followed up with manual MICs and further organisms in the Warwick Antimicrobial Screening Facility. Our expert
scientists are always on hand to help with data analysis and investigation, to make sure you get the as much from your data as possible.
