Bottom-up Decoding of Protein Conformational Landscapes: from Gas-phase to Solution
This is a fully-funded 4-year PhD position based in the HetSys Centre for Doctoral Training at the University of Warwick.
Project outline
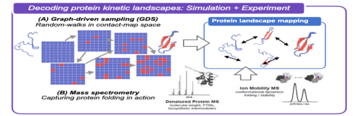
Protein folding is a dynamic, complex process by which proteins traverse the free energy landscape, thus connecting the unfolded and the folded (native) state. Understanding protein folding mechanisms is a key route to better understanding misfolding-related diseases such as Alzheimer's - but predicting how proteins fold in biological environments remains a key unmet challenge.
This project brings together insights from efficient graph-driven folding simulations with mass spectrometry experimental data, creating a unique multi-stranded methodology to map out free energy landscapes associated with protein folding in environments spanning gas-phase to microsolvation environment.
Supervisors
Primary: Prof. Scott Habershon (Chemistry)
Dr Michael Faulkner (Engineering)
The overall aim of the project is to develop an efficient computational strategy for high-throughput mapping of the free energy landscapes associated with protein-folding, and to subsequently use this approach to interpret the results of biological mass spectrometry experiments performed on a range of different proteins of practical interest. Specific project milestones include:
- Development of new efficient machine-learning (ML) strategies based on graph neural networks - to predict relative free energies of different protein contact-map states in graph-driven sampling;
- Design simulation protocols for evaluation of kinetic transition rates between protein-folding intermediates;
- Deploying novel ML-driven graph-based sampling to map protein-folding landscapes for target proteins identified in collaboration with experimental researchers;
- Cross comparison of predicted protein-folding paths with mass spectrometry experimental data, picking apart the role of sequence.
This project will deliver a new computational approach - uniquely validated, informed and characterized with state-of-the-art mass spectroscopy experimental data provided by experimental colleague Dr. Matt Jenner in the Department of Chemistry to disentangle protein folding/unfolding pathways in systems ranging from gas-phase to microsolvation.
This collaborative effort is a unique strength of the project andprovides a prime opportunity to study how protein folding is coupled to the solvent environment - through the lens of both theory and experiment.
This is an important goal - for example, protein mis-folding is associated with a range of diseases such as Alzheimer's and cystic fibrosis, hence new insights into folding and mis-folding mechanisms is essential to support biological insights. We aim to release associated software via GitHub, once sufficient test-cases, documentation and robustness is established. Based on previous experience, we anticipate 2-3 journal articles.
Students will develop skills in:
Scientific software development using Python and Fortran;
Robust software development practices, such as version control and automated documentation;
High-performance computing strategies, delivered through formal HetSys training and application throughout the project;
Writing and presentation, through delivery of journal articles and conference presentations disseminating the outcomes of the project;
Machine-learning techniques, through implementation of new graph neural network models for free energy prediction;
Computational chemistry and molecular simulation expertise;
Teamwork and collaboration, through direct engagement with experimental researchers at Warwick, primarily the research team of Dr. Matthew Jenner (Chemistry).
These skills position you for careers in AI research, computational materials science, national laboratories, tech industry or academic research. The HetSys training provides a foundation for these skills through dedicated courses and cohort activities.
Generating Protein Folding Trajectories Using Contact-Map-Driven Directed Walks; Ziad Fakhoury, Gabriele C. Sosso, Scott Habershon; J. Chem. Inf. Model. 2023, 63, 7, 2181–2195; https://doi.org/10.1021/acs.jcim.3c00023
Contact-Map-Driven Exploration of Heterogeneous Protein-Folding Paths; Ziad Fakhoury, Gabriele C. Sosso, Scott Habershon; J. Chem. Theory Comput. 2024, 20, 18, 8340–8353; https://doi.org/10.1021/acs.jctc.4c00878
We require at least a II(i) honours degree at BSc or an integrated masters degree (e.g. MPhys, MChem, MSci, MEng etc.) in a physical sciences, mathematics or engineering discipline. We do not accept applications from existing PhD holders.
If you are an overseas candidate please check here that you hold the equivalent grades before applying.
For postgraduate study in HetSys, the term “overseas” or “international” student refers to anyone who does not qualify for UK home fee status. This includes applicants from the European Union (EU), European Economic Area (EEA), and Switzerland, unless they hold settled or pre-settled status under the UK’s EU Settlement Scheme.
If you are a European applicant without UK residency or immigration status that qualifies you for home fees, you will be classified as an overseas student.


