Michael Stevens
I am currently doing a PhD at the university of Dundee under the supervision of Dr David Norman.
Project
My project involves trying to advance EPR as a technique for determining the structure of large biological molecules. Increasing the number of restraints one can obtain from a spin label pair, on a biological structure, is one way of doing this. A spin label is a stable radical which can be introduced into the desired biological molecule at a specific site. Currently the most popular spin label used in proteins is the nitroxide radical containing MTSL, which forms a disulphide bond with cysteine to make the R1 residue. To spin label a protein with MTSL, site directed mutagenesis is used, to firstly remove unwanted cysteine residues from the structure, followed by the introduction of cysteine at sites where the label is required. In normal practice distance distributions between spin labels are determined using Pulsed Electron-Electron Double Resonance (PELDOR).
PELDOR uses pulses of microwave radiation set at different frequencies to excite different groups of spin labels. Spin labels close enough to exhibit dipolar coupling produce signals of different strength based upon the timing between the two pulses. Due to the increased g-tensor at high frequencies, such as W-Band, nitroxide radicals can be selectively excited based upon their orientation to the external magnetic field. This allows PELDOR select groups of spin labels based upon their orientation to the magnetic field and thus determine the orientation between the spin label pair. Though the rotation around bonds of the R1 side chain is restricted, it is still too mobile for any orientation information from the spin label to be translated into structural information for the protein. Rx (below) is a bi-functional spin label based upon R1, the second leg of the spin label further restrains its motion. This makes it possible for orientations between spin labels to be turned into useful structural information for the bound protein.
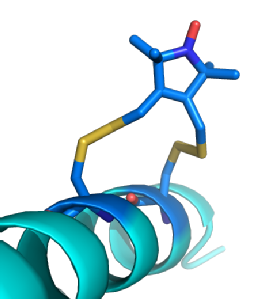
(Left) A model of Rx (Dark Blue) bound to an Alpha helix. Rx can bind to the Alpha helix and Beta sheet secondary structures. These provide stable platforms where variations in distance and/ or orientations are caused almost entirely by the spin label. This makes the spin labels behaviour anisotropic allowing the position of the protein to be modelled based upon measurements from the nitroxide radical.
Another method of improving structural information gained upon large biomolecules by EPR is to change the biophysical properties of the sample. One of the limitations to the acquisition of PELDOR data is the de-phasing of relaxing electrons by a number of mechanisms. In the solid state, the de-phasing time is known as Tm and in biological samples the main source of reduction in Tm is surrounding protons. To increase the Tm it is now common practice to run samples in deuterated solvents and recently a study has been done using a deuterated protein showing a further large increase in Tm. Because the sensitivity of PELDOR depends on the size of the measured spin-echo, increasing Tm raises increases sensitivity of PELDOR. The persistence of the spin-echo also determines the maximum distance between spin-labels that can be measured. More recently the affect of protein deuteration upon the Tm has been further investigated by selective deuteration of the different proteins componants in the nucleosome core octomer (unpublished data). This study demonstrated the distance dependence effect on Tm (lone electron to proton). This affect could be further investigated by the introduction of individually protonated amino acids into a deuterated protein matrix.

Contact Details:
e-Mail: m.a.stevens@dundee.ac.uk
Phone Number: +44(0)1382 388712