Alternative protein sources: growing the next generation computational modelling framework
Supervisors: Radu Cimpeanu and Ferran Brosa Planella
Summary:
The alternative protein space represents one of the most dynamic scientific areas at present. Shrinking usable land mass, even accounting for agricultural advances, means a sustainable future is tightly linked with our ability to create and support clean food sources. Computational modelling has an immense (and largely untapped) potential to innovate a technological space still in its infancy. We will use a combination of state-of-the-art techniques - multi-physics fluid mechanics, high performance computing and data-driven approaches - to design a versatile open-source modelling framework supported by fantastic experimental and industrial collaborators from around the world.
Background:
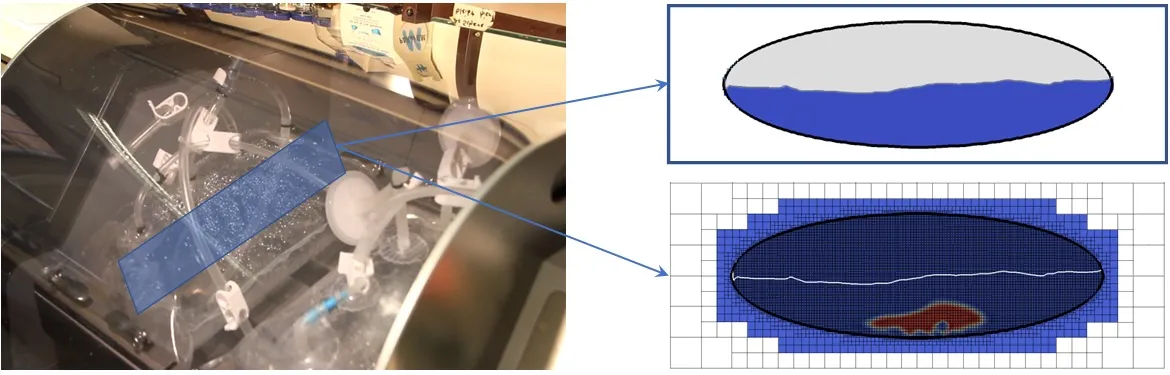
The primary objective of the project is to facilitate sustained progress within a new and rapidly evolving landscape – the science behind cultivated meat, which is only just starting to benefit from research at the interface between continuum modelling and high performance computing. As a concrete vehicle for innovation, rocking wave bioreactors (shown below) have been identified as a key technology given the low shear forces and improved gas exchange at the gas-liquid interface [2]. The feasibility and potential impact of improving bioreactors show excellent promise [1], while the knowledge base so far has been primarily built outside the field of fluid mechanics, relying on oversimplifying assumptions [3], expensive and non-generalisable black-box software [4], or trial-and-error efforts in practical contexts. This project will thus support a modelling stream which has only been identified within the last few years, promoting a rigorous open scientific interdisciplinary culture in this area for the first time.
The initiative will be accelerated via recently acquired expertise and rapidly expanding international collaborative and industrial networks, with Prof. Daniel Harris at Brown, an enthusiastic academic collaborator with modelling and experimental capabilities, and scoping work performed with partners ranging from start-ups to corporations via the Good Food Institute and the Cultivated Meat Modelling Consortium). At present there are two enthusiastic students and one post-doctoral researcher in the extended group (Warwick + Brown) working on this topic. There are also opportunities to link up with local initiatives at Warwick such as our very own Warwick Food Global Research Priority program.
Links to HetSys Training
The project, which lies at the interface between subfields in mathematics and engineering (fluid mechanics, bioengineering, agent-based modelling), is ideally suited for the HetSys training and research ethos. Modules such as PX911 (multi- scale modelling), PX912 (mechanics), PX913 (advanced computational skills), PX914 (uncertainty quantification), have a near-perfect translation into the project methodology, with optional modules PX921 and PX923 acting as specialisation routes, all in a supportive, collaborative cohort environment.
Whilst fundamental in nature, the project brings together elements from diverse areas in modern applied mathematics, biochemistry and software engineering. The student will draw extensive knowledge from the initial training year, whilst benefitting from the expertise of each of the supervisors and external partners to guide the project-specific skillset development.
The biological nature of the subject matter (including phenomena such as phase change and rheology, as well as operational aspects) brings excellent opportunities to include uncertainty quantification into the modelling effort.
The project incorporates a strong software engineering component. There are already established contributions in our group in the implementation and testing of direct numerical simulation capabilities using high quality and well maintained open-source software (e.g. Basilisk and oomph-lib), including dissemination of tutorials and benchmark data.
References
[1] Humbird, Biotech. and Bioeng. 118, 2021.
[2] Junne et al., Chem. Ing. Tech. 85, 2013.
[3] Westbrook et al., Biochem. Eng. J. 88, 2014.
[4] Zhan et al., Chem. Eng. Sci. 193, 2019.
