Linkage Maps
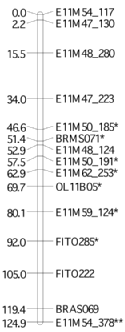
When all the members of the population have been scored (genotyped) with a set of molecular markers, the data can be used to make a linkage map (often described as a genetic map). The linkage map describes the linear order of markers within linkage groups.
This process estimates how the markers are grouped together based on the number of recombination breaks between them (recombination frequency) with one genetic map unit equal to the distance between marker pairs for which one product of meiosis out of a hundred is recombinant (recombination frequency of 1 per cent).
A mapping function (Haldane, Kosambi or Morgan) converts the recombination frequencies into mapping units, typically centimorgan units.
If we can resolve the genotype data into the same number of linkage groups as there are chromosomes we can use markers within groups that have previously been shown to map to a particular chromosome to order our linkage groups relative to their correct chromosomal assignments.
 |
|
The linkage map is an essential tool for research on plants whose genomes have yet to be sequenced, since it provides a framework of marker order and spacing. This in-turn allows comparative analyses with the maps and sequence of other species. The linkage map also serves as a starting point to map quantitative trait loci (QTL) for target traits. For example broccoli head morphology and leaf shape.
In the age of complete genome sequence assembled into pseudo-chromosomes, some might say that the linkage group has 'had its day' - this is far from true. The linkage map still provides an essential backbone for sequence assembly and validation. Indeed the estimation of QTL locations on a linkage map is still needed to bridge from markers to candidate gene models embedded in the assembled sequence data. The linkage map then provides information on linked markers that can be used during marker-assisted selection in breeding programs, making the selection process more efficient.
