Blog
How to do the codon-optimisation using Boost JGI website
Before starting, you have to register for the boost website here. It's free.
1. go to https://boost.jgi.doe.gov/boost.html#Link opens in a new window and choose polish

2. select the polish option, enter/upload the sequence of the gene of interest and specify constraints. You can click on Gen9 or JGI on the right hand and click validate.
To put your sequence in fasta format, don't forget to put >gene in front of the sequence. For example, if my sequence is;
atgacgttgtcggtcgtggtgctcgtcgggtcctcggtggaccgctttcacagtgatctc
tctcgggtctacgccggcggcttcctgggtgccctcggtaacgaccccgcgtacggtttc
gagatcgcgttcgtctctcccggcggcctctggagtttccccaccgatctcgaggattcg
I'll have to put the mysD name at the beginning like;
>mysD
atgacgttgtcggtcgtggtgctcgtcgggtcctcggtggaccgctttcacagtgatctc
tctcgggtctacgccggcggcttcctgggtgccctcggtaacgaccccgcgtacggtttc
gagatcgcgttcgtctctcccggcggcctctggagtttccccaccgatctcgaggattcg

3. Select a modification strategy. I chose the balance option. Choose the codon usage for the organisms that you will put your gene into. Streptomyces is not on the list, so, I used the codon usage for Streptomyces coelicolor from this >> kazusa website instead. Just copy and paste.
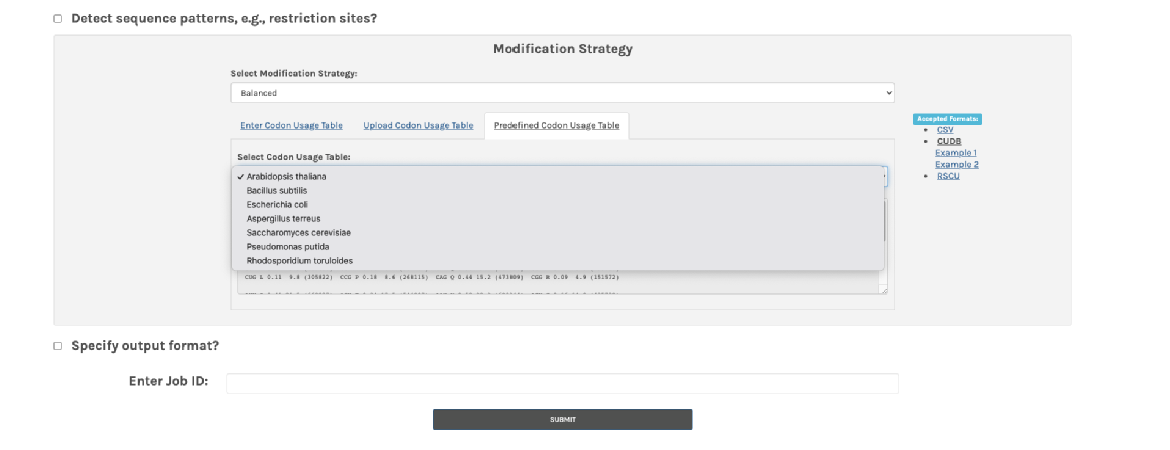
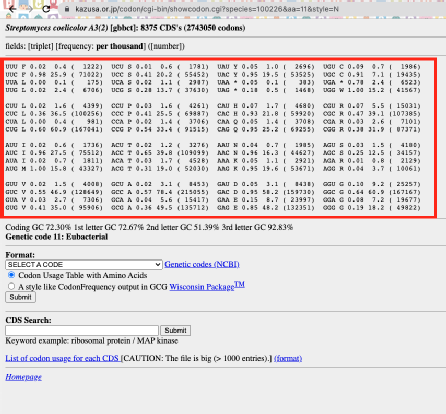
Copy and paste the red box into the Enter codon usage table on the boost website. Click submit and wait a few seconds to see the results.
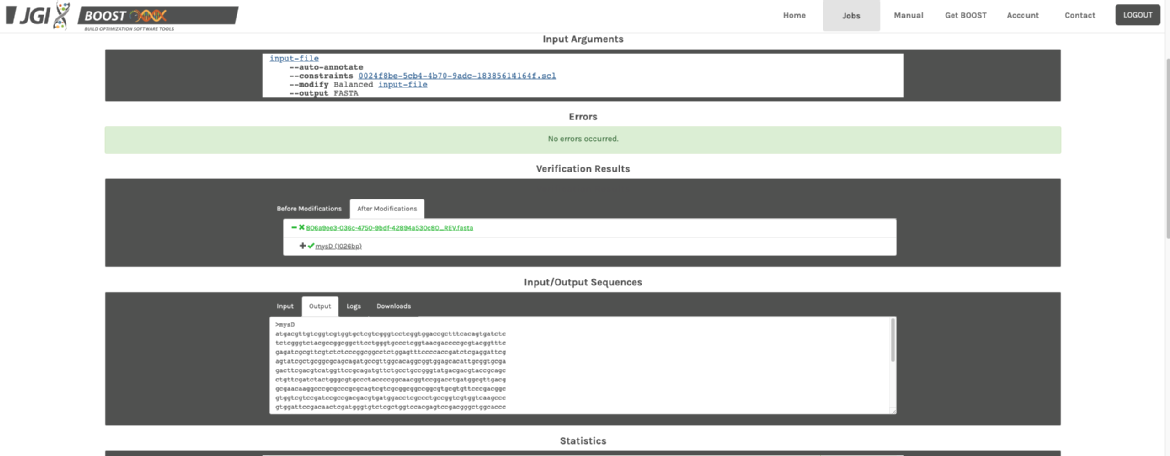
4. You can either download the results or copy the output sequence directly. The sequence is already codon-optimised! 🎉