Chrystala Constantinidou
Summary
Pathogenic bacteria: Regulation, assembly and function of macromolecular structures used for motility and secretion
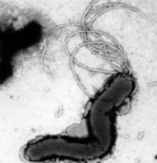
Biogenesis of polar flagella
The flagellum is a core virulence determinant for a number of pathogenic bacteria, important not only in motility and chemotaxis but also in colonisation, host invasion, biofilm formation and protein secretion. Flagella-gene expression is a hierarchical process, with layers of exquisite and tightly controlled regulation that still intrigue and surprise. Different bacteria species are characterised by diverse patterns and numbers of flagella, with each genome carrying different panoply of flagella-associated genes. We are focusing our studies on bacteria such as Campylobacter jejuni and Helicobacter pylori, trying to understand which genes and patterns of expression determine the different flagella patterns observed in these two closely related bacterial species.
Secretion Systems
Bacteria employ a wide variety of protein secretion systems that enable them to translocate substrates including small molecules, proteins and DNA outside of their cells and sometimes into neighbouring prokaryotic or eukaryotic cells. These substrates play key roles allowing the bacteria not only to interact and modify their immediate environment but also contribute to physiological process such as adhesion, pathogenicity, adaptation and survival. Secretion requires translocation across the inner membrane, the outer membrane for Gram-negative bacteria and across dense envelopes, as is the case with mycobacteria. We are studying, a secretion system never before described in Gram-negative bacteria and our initial studies are focused on the function and role of such a system in Helicobacter pylori.
Small RNAs
Non-coding RNAs (ncRNAs) have been shown to perform a wide range of regulatory functions, important not only for bacterial homeostasis but also playing significant roles in bacterial virulence. We are interested in a particular class of bacterial ncRNAs, small regulatory RNAs (sRNAs), which regulate trans-encoded mRNAs at the post-transcriptional level.
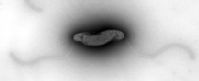 In the Gram-negative model organism Escherichia coli, the function of sRNA’s depends on the presence of the RNA binding protein Hfq, which sits at the core of the post-transcriptional regulatory network in this species. However, Hfq does not perform this role in all species and many bacterial species lack a homologue of Hfq altogether, including the important human pathogen Campylobacter jejuni.
In the Gram-negative model organism Escherichia coli, the function of sRNA’s depends on the presence of the RNA binding protein Hfq, which sits at the core of the post-transcriptional regulatory network in this species. However, Hfq does not perform this role in all species and many bacterial species lack a homologue of Hfq altogether, including the important human pathogen Campylobacter jejuni.
Through the use of “omics” technologies (such as transcriptomics and proteomics) combined with molecular biology and novel global RNA-RNA interaction studies we are trying to define and understand how C. jejuni utilise sRNA’s and the factors involved in this network.
Bacterial genome sequencing and microbiome studies
We are involved in a variety of projects, clinical and environmental, all utilising the ever-increasing power of high throughput sequencing. For example:
• Sequencing bacterial species associated with Cystic Fibrosis patients
• Analysing the cervical microbiome of mothers with pre-term babies
• Exploring longitudinal changes in the cervical microbiome
• Utilising 16S and metagenomic analysis to understand how environmental microbiomes are affected by agriculture.
Selected publications:
Loman NJ, Constantinidou C, Christner M, Rohde H, Chan JZ, Quick J, Smith GP, Betley JR, Weir JC, Aepfelbacher M, Pallen MJ (2013) Diagnostic Metagenomics: A Culture-Independent Approach to the Diagnosis Of Bacterial Infection Using High-Throughput Sequencing. JAMA 309, 1502-1510
Loman NJ, Constantinidou C, Chan J, Halachev M, Sergeant M, Penn CW, Robinson ER, Pallen MJ (2012) High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nature Reviews Microbiology 10: 599-606
Kamal N, Dorrell N, Jagannathan A, Turner SM, Constantinidou C, Studholme DJ, Marsden G, Hinds J, Laing KG, Wren BW, Penn CW (2007) Deletion of a previously uncharacterized flagellar-hook-length control gene fliK modulates the σ54-dependent regulon in Campylobacter jejuni. Microbiology 153(9):3099-111
Zheng D, Constantinidou C, Hobman JL, Minchin SD (2004) Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Research 32: 5874-5893