Anisha Haris
PhD Project
To utilize the electron-based fragmentation and photodissociation methods available on the Bruker solariX 12T FT-ICR MS to differentiate isomers of amino acids in peptides and proteins.
Current work:
1. Differentiation and quantitation of the isomeric products of deamidation in peptides using ECD MS/MS.
The spontaneous, non-enzymatic deamidation of asparagine (Asn) (and to a lesser extent glutamine, Gln) is a common PTM, playing a significant role in diseases related to ageing such as Alzheimer’s and Parkinson’s disease.
The ability to distinguish between the isomerization products of Asn deamidation and the quantitation of the ratio of these isomers will help us to gain a deeper understanding of the effect that they have on protein structure, which is known to alter the activity and function of the proteins. For example, in monoclonal antibodies, the isomerisation of Asp to isoAsp reduces the antigen binding affinity and thereby reduces the overall antibody activity.
Chromatography and tandem MS methods such as collisionally activated dissociation can be used to separate Asp from isoAsp but it can be challenging due to the slight structural differences between the isomers. Hence using ultra-high resolution FT-ICR MS and employing electron based fragmentation methods may be more beneficial as we can generate diagnostic fragment ions for Asp and isoAsp and quantify the ratio of these isomers using electron capture dissociation (ECD).
Cleavage of the Cα - Cβ bonds in the peptide backbone by ECD MS/MS generates diagnostic fragment ions for isoAsp residues, cl-n + 57 and zn—57.

Figure 1. a) deamidation of Asn and isomerization of Asp to isoAsp via hydrolysis of the succinimide intermediate and b) deamidation of Gln.
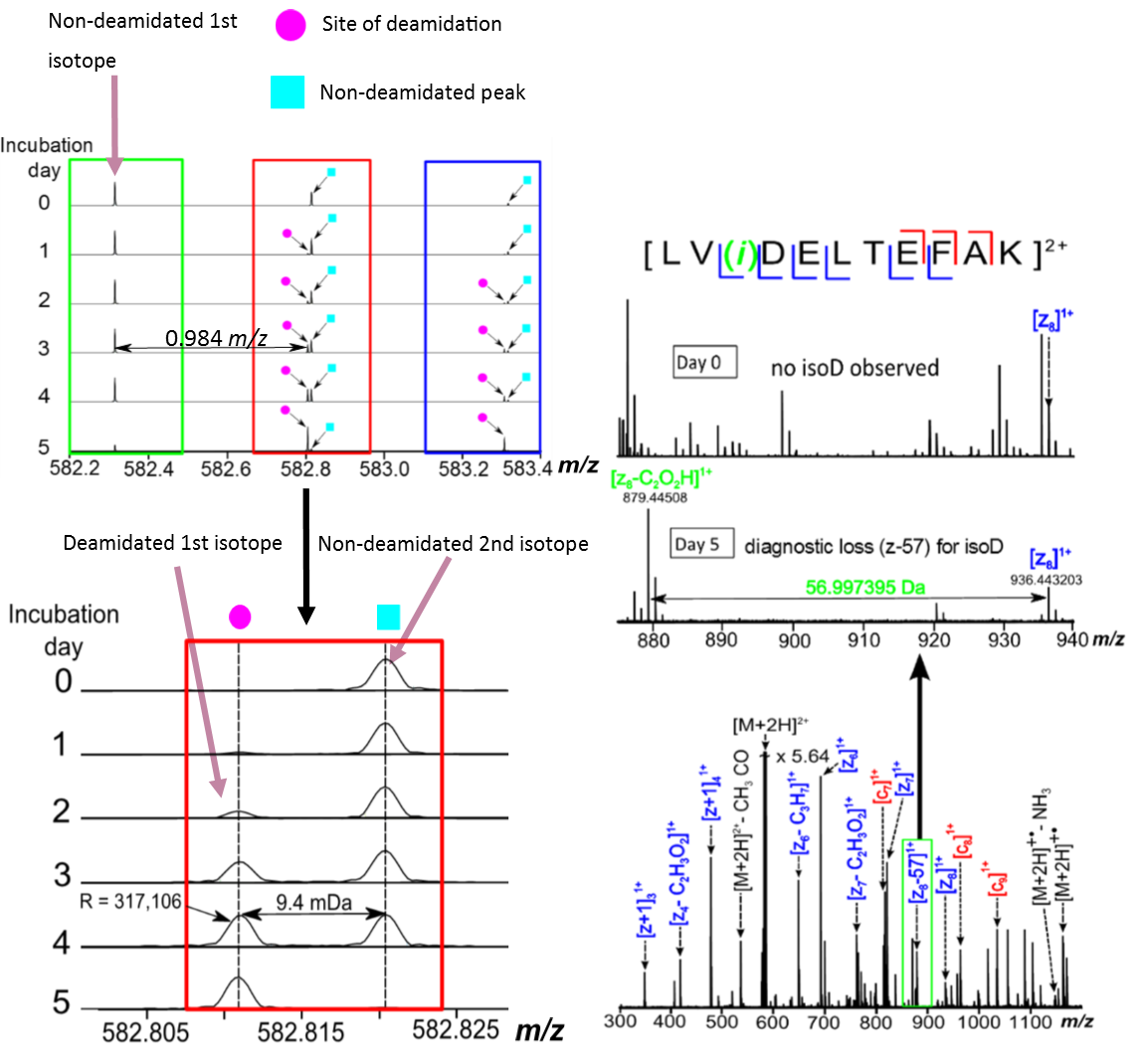
Figure 2. MS and ECD MS/MS spectra of a bovine serum albumin (BSA) trypsin digested deamidated peptide.
2. Distinguishing between methylated histidine isomers generated as a post-translational modification of actin
This is a collaborative project with the division of Biomedical Sciences (cell development and biology programme) at Warwick Medical School. 3-methylhistidine (3-MH) is formed by methylation of histidine as a post-translational modification of the muscle and motor proteins, actin and myosin. 3-MH is of particular interest as it is liberated during degradation of these proteins and therefore can be used a biomarker for muscle degeneration when found in urine or plasma.
Figure 3. Chemical structures of the methylhistidine isomers, 1-MH and 3-MH.
3. Exploring tandem mass spectrometry methods for the analysis of dihydroxylated vitamin D3 isomers
Vitamin D compounds are a group of fat-soluble secosteroids, which are vital for maintaining bone health in humans. In particular, recent studies have shown that the dihydroxylated vitamin D3compounds: 1,25-dihydroxyvitamin D3(active form of vitamin D) and 24,25-dihydroxyvitamin D3 (inactive form) have significant biological effects, playing a role in diseases such as osteoporosis.
Figure 4. Metabolic pathway of Vitamin D.
Identification and differentiation of the isomers by mass spectrometry can be challenging due to the zero mass difference and minor structural differences between them. The isomers may require separation by liquid chromatography (LC) with the presence of a derivitization agent, which can add extra complexity to the spectra.
Here, we investigated the use of fragmentation methods, particularly electron induced dissociation (EID) and ultraviolet photodissociation (UVPD), which are available on a Bruker 12T Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR MS). EID uses high energy electrons generated from a hollow dispenser cathode to fragment the ions. UVPD is implemented using a 193 nm ArF excimer laser, which has shown to result in extensive fragmentation of peptides, proteins, lipids and other small molecules.
Master’s Project
Application of Differential Ion Mobility Spectrometry-Mass Spectrometry to the Analysis of Isomeric Lipids.
Academic Background
Completed an integrated Master’s degree (MChem) at the University of Bradford.
Degree of Bachelor of Science with First Class Honours in Chemistry (2013-2016)
Master in Chemistry with Distinction in Chemistry with Industrial Experience (2016-2017)
Industrial Placement
Shimadzu Research Laboratory
Manchester, UK
During my Master’s, I carried out a 12-month extended placement at Shimadzu Research Laboratory (SRL) . At SRL, I was primarily involved in applications work testing an advanced prototype mass spectrometer incorporating a differential ion mobility spectrometer (DMS). I was able to gain significant hands on experience with single and triple quadrupole mass spectrometers, time-of flight, as well as the DMS. Whilst primarily applications-based, I was also involved with debugging and maintaining the instrumentation where necessary.
Publications
Shvartsburg A.A, Haris A, Andrzejewski R, Entwistle A, Giles R. Differential Ion Mobility Separations in the Low-Pressure Regime. Analytical Chemistry, 2017.
DOI: 10.1021/acs.analchem.7b03925
Oral Presentations
Anisha Haris, Yuko P.Y. Lam, Christopher A. Wootton, Alina Theisen, Tomas E. Morgan, Cookson K.C. Chiu, Mark P. Barrow and Peter B. O'Connor. Tandem Mass Spectrometry for Isomer Differentiation. August 2019. 4th Short Course EU FT-ICR MS Network - University of Warwick
Anisha Haris, Yuko P.Y. Lam, Christopher A. Wootton, Alina Theisen, Tomas E. Morgan, Cookson K.C. Chiu, Mark P. Barrow and Peter B. O'Connor. Differentiation and Quantitation of the Isomeric Products of Deamidation using FT-ICR MS. October 2019. East Midlands Proteomics Workshop (EMPW). Sheffield, UK.
Anisha Haris, Yuko P.Y. Lam, Christopher A. Wootton, Alina Theisen, Tomas E. Morgan, Cookson K.C. Chiu, Mark P. Barrow and Peter B. O'Connor. Differentiation and Quantitation of the Isomeric Products of Deamidation using FT-ICR MS. May 2020. Department of Chemistry Virtual Postgraduate Symposium. University of Warwick, UK.
Poster Presentations
Anisha Haris, Yuko P.Y. Lam, Tomas E. Morgan, Cookson K.C. Chiu, Christopher A. Wootton, Mark P. Barrow and Peter B. O'Connor. Quantitating the Ratio of Isoaspartic Acid to Aspartic Acid in Proteins by Electron Capture Dissociation. July 2018. Uppsala Conference on Electron Capture and Transfer Dissociation Mass Spectrometry (Uppcon) in Leeds, UK.
Anisha Haris, Yuko P.Y. Lam, Tomas E. Morgan, Cookson K.C. Chiu, Christopher A. Wootton, Mark P. Barrow and Peter B. O'Connor. Buffer Concentration Effects on the Isomerization Products of Deamidation in Peptides using ECD MS/MS. September 2018. BMSS in Cambridge, UK.
Anisha Haris, Yuko P.Y. Lam, Christopher A. Wootton, Alina Theisen, Tomas E. Morgan, Cookson K.C. Chiu, Mark P. Barrow and Peter B. O'Connor. Comparison of ECD and UVPD to Distinguish between the Isomeric Products of Deamidation using FT-ICR MS. March 2019. Celebration of Native Mass Spectrometry in Oxford, UK.
Anisha Haris, Yuko P.Y. Lam, Christopher A. Wootton, Alina Theisen, Tomas E. Morgan, Cookson K.C. Chiu, Mark P. Barrow and Peter B. O'Connor. Differentiation and Quantitation of the Isomeric Products of Deamidation using FT-ICR MS. September 2019. BMSS in Manchester, UK.
Anisha Haris, Yuko P.Y. Lam, Alina Theisen, Christopher A. Wootton, Bryan P. Marzullo, Pascal Schorr, Yulin Qi, Dietrich Volmer, and Peter B. O'Connor. Differentiation of Dihydroxylated Vitamin D3 Isomers using Tandem Mass Spectrometry. March 2020. DGMS in Münster, Germany.
Anisha Haris, Yuko P. Y. Lam, Alina Theisen, Christopher A. Wootton, Tomos E. Morgan, Mark P. Barrow, and Peter B. O’Connor. Comparison of ECD and UVPD for the Relative Quantitation of the Isomeric Products of Deamidation. June 2020. ASMS Online Reboot Program.
Conferences
British Mass Spectrometry Society (BMSS), September 2017 – Manchester, UK
Uppcon, July 2018 - Leeds, UK
EU FT-ICR MS Summer School, August 2018 - Joensuu, Finland
BMSS, September 2018 - Cambridge, UK
East Midlands Proteomics Workshop (EMPW), October 2018 - Lincoln, UK
Celebration of Native Mass Spectrometry, March 2019 - Oxford, UK
4th Short Course EU FT-ICR MS Network, August 2019 - University of Warwick, UK
BMSS, September 2019 - Manchester, UK
18th EMPW, October 2019 - Sheffield, UK
Awards
July 2017 – Awarded the JEOL UK ltd. prize for best overall performance in Stage 4 of an MChem course
September 2018 - BMSS John Beynon Travel Award for BMSS Conference in Cambridge, UK
October 2019 - Awarded for the best oral presentation at EMPW in Sheffield, UK
March 2020 - BMSS John Beynon Travel Award for DGMS Conference in Münster, Germany
FT-ICR MS Group
Department of Chemistry
The University of Warwick
Millburn Hill Road
Coventry
CV4 7AL
Office: +44 (0) 24 7615 1304
Lab: +44 (0) 24 7615 1305
Email: A.Haris@warwick.ac.uk
