Alexiane Decout
Summary
The vaginal microbiome is a complex environment playing a key role in women’s reproductive health. Vaginal dysbiosis, leading to inflammation of the lower female reproductive tract, is associated with a broad range of women’s reproductive health disorders such as bacterial vaginosis, increased susceptibility to sexually transmitted infections and pregnancy complications like preterm birth and infertility, impacting millions of women and babies each year in the UK alone.
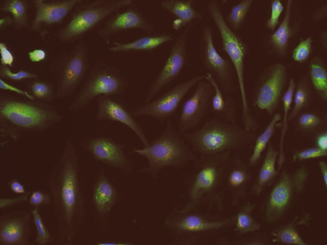 We are interested in understanding how the vaginal microbiota modulates the maternal innate immune environment to promote health or disease. We previously showed that Lactobacillus crispatus, commensal bacterium associated with optimal vaginal health, selectively interacts with the anti-inflammatory innate immune receptors. We also identified the Surface Layer Proteins (SLPs) of Lactobacillus crispatus as key immunoregulatory molecules in the context of healthy microbiota.
We are interested in understanding how the vaginal microbiota modulates the maternal innate immune environment to promote health or disease. We previously showed that Lactobacillus crispatus, commensal bacterium associated with optimal vaginal health, selectively interacts with the anti-inflammatory innate immune receptors. We also identified the Surface Layer Proteins (SLPs) of Lactobacillus crispatus as key immunoregulatory molecules in the context of healthy microbiota.
Our aim is to comprehensively understand the interplay between the maternal innate immune system and the vaginal microbiota. To do so, we use a combination of reporter cells, primary cells and patient samples to identify i) the maternal innate immune receptors regulating the immune environment and ii) the immunomodulatory molecules produced by the vaginal microbiota and detected by the maternal immune system. Our work will inform the development of vaginal live biotherapeutic products with optimal innate immune properties, as well as host-targeted immunomodulatory therapeutics for the prevention and treatment of adverse women’s health outcomes.
Selected publications:
Yu J., Decout A., Di Maio A., Chai W., Stehle T., Turnbull B., Feizi T., Liu Y., Novel multifunctional glycan probes to elucidate specificities of diverse glycan recognition systems, biorxiv.org/cgi/content/short/2023.07.09.548266v1, (Preprint)
Decout A., Katz J. D., Vankatraman S., Ablasser A. (2021) The cGAS–STING pathway as a therapeutic target in inflammatory diseases, Nature Reviews Immunology, 21, pages 548–569,
Pathare G. R.*, Decout A.*, Glück S., Cavadini S., Makasheva K., Hovius R., Kempf G., Weiss J.,Kozicka Z., Guey B., Melenec P., Fierz B., Thomä N. H., Ablasser A. (2020) Structural mechanism of cGAS inhibition by the nucleosome, Nature, 587:668-672.
Guey B.*, Wischnewski M.*, Decout A., Makasheva K., Kaynak M., Sakar M. S., Fierz B., Ablasser A. (2020) BAF restricts cGAS on nuclear DNA to prevent innate immune activation, Science, 369: 823-828
Decout A., Silva-Gomes S. et al. (2018) Molecular bases of mycobacteria and lipoglycan recognition by the C-type lectin Dectin-2, Sci. Rep., 8 (1): 16840
Decout A & Ablasser A (2019) Virology: poxin soothe the STING, Current Biology, 29 (9): R332-R334
Decout A & Ablasser A (2018) Human cGAS has a slightly different taste for dsDNA, Immunity, 49 (2):206-208
Haag S. M., Gulen M. F., Reymond L., Gibelin A., Abrami L., Decout A., Heymann M., van der Goot G.F., Turcatti G., Behrendt R., Ablasser A. (2018) Targetting STING with covalent small molecule inhibitors, Nature, 559: 269-273
Decout A., Silva-Gomes S., Drocourt D., Barbe S., André I., Cueto F. J., Lioux T., Sancho D., Pérouzel E., Vercellone A., Prandi J., Gilleron M., Tiraby G., Nigou J. (2017) Rational design of new adjuvants targeting the C-type lectin Mincle, Proc. Natl. Acad. Sci., 114 (10), 2675-2680
Silva-Gomes S., Decout A., Nigou J., Pathogen-associated molecular patterns. In: Parnham BSc, PhD, CBiol MSB, FBPharmacolS M. (Ed.) Encyclopedia of Inflammatory Diseases: SpringerReference (www.springerreference.com). Springer-Verlag Berlin Heidelberg, 2014

