Publications
A complete up-to-date list of publications of Robert can be found on Google Scholar and full-texts might be available from the Warwick Research Active Portal (WRAP).
Zinc finger homeobox-3 (ZFHX3) orchestrates genome-wide daily gene expression in the suprachiasmatic nucleus
 |
New preprint from Pat Nolan's lab by Akaknsha Bafna looks at the contribution of the ZFHX3 deletion in adult mice for SCN transcriptome and cistrome. Vadim contributed TimeTeller analysis of SCN data suggesting a significant phase advance in entrained SCN of mutant mice. |
Adenosine Kinase regulates Sleep Timing and the Homeostatic Sleep Response through Distinct Molecular Pathways
A follow up on the actions of adenosine kinase in collaboration with Aarti Jagannath's group.
Sleep behaviour is broadly regulated by two drives, the circadian (Process C), which is orchestrated by the suprachiasmatic nuclei (SCN), and controls sleep timing, and the homeostatic (Process S), which controls sleep amount and the response to sleep deprivation (Borbély et al., 2016). However, the molecular pathways that mediate their independent effects, and their interactions remain unclear. Adenosine is an important integrator of both processes (Bjorness & Greene, 2009; Jagannath et al., 2021, 2022), such that adenosine levels track and modulate wakefulness, whilst adenosine signalling inhibits the circadian response to light. Therefore, we studied the sleep/circadian behaviour, and cortical and SCN transcriptomic profiles of a mouse model overexpressing Adenosine Kinase (Adk-Tg) (Fedele et al., 2005), (Palchykova et al., 2010). We found that overall, the Adk-Tg mouse slept less and showed lower amplitude circadian rhythms with an altered sleep/wake distribution across the 24h day, which correlated with changes in transcription of synaptic signalling genes that would shift the excitatory/inhibitory balance. In addition, the Adk-Tg mouse showed a reduced level of ERK phosphorylation, and attenuation of DNA repair related pathways. After sleep deprivation, however, the Adk-Tg mouse significantly increased relative to wildtype, immediate early gene expression levels including of Arc, but paradoxically reduced ERK phosphorylation. Thus, baseline sleep levels and timing are regulated by ERK signalling, whereas the response to sleep loss is mediated by the alteration of the transcriptomic landscape independently of ERK.
TimeTeller: a tool to probe the circadian clock as a multigene dynamical system
|
|
Great collaboration started by David Rand and Francis Lévi with contributions from many including MRC DTP PhD students Laura Usselmann and Vadim Vasilyev, we describe a novel tool to interrogate the circadian clock from a single sample's transcriptome. We show in many examples how this can deliver useful information not only on the phase of the biological clock in the sample, but also give an estimate on the functionality of the clock. Furthermore, we show how this has potential as a biomarker to stratify data-sets from human tissues as well as inform research in experimental models. The TimeTeller algorithm will be available for use to any interested colleague soon. |
Flow Rate Independent Multiscale Liquid Biopsy for Precision Oncology
Collaborative work with Jerome Charmet and Holosensor Medical Technology Ltd.
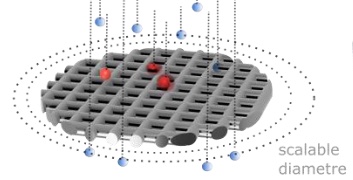 |
Immunoaffinity-based liquid biopsies of circulating tumour cells (CTCs) hold great promise for cancer management, but typically suffer from low throughput, relative complexity and post-processing limitations. Here we address these issues simultaneously by decoupling and independently optimising the nano-, micro- and macro-scales of a CTC enrichment device that is both simple to fabricate and operate. Our device achieved an 80% positive match in the identification of HER2+ breast cancer (n=26) compared to clinical standard FISH on solid biopsy. The results suggest that our approach, which overcomes major limitations previously associated with affinity-based liquid biopsies, could provide a versatile tool to improve cancer management. |
First Preprint: Cell-type specific circadian bioluminescence rhythms recorded from Dbp reporter mice reveal circadian oscillator misalignment
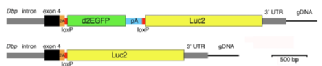 |
Circadian rhythms are endogenously generated physiological and molecular rhythms with a cycle length of about 24 h. Bioluminescent reporters have been exceptionally useful for studying circadian rhythms in numerous species. Here, we report development of a reporter mouse generated by modification of a widely expressed and highly rhythmic gene encoding D-site albumin promoter binding protein (Dbp). In this line of mice, firefly luciferase is expressed from the Dbp locus in a Cre-recombinase-dependent manner, allowing assessment of bioluminescence rhythms in specific cellular populations. A mouse line in which luciferase expression was Cre-independent was also generated. The Dbp reporter alleles do not alter Dbp gene expression rhythms in liver or circadian locomotor activity rhythms. In vitro and In vivo studies show the utility of the reporter alleles for monitoring rhythmicity. Our studies reveal cell-type specific characteristics of rhythms among neuronal populations within the suprachiasmatic nuclei in vitro. In vivo studies show stable Dbp-driven bioluminescence rhythms in the liver of Albumin-Cre;DbpKI/+ liver reporter mice. After a shift of the lighting schedule, locomotor activity achieved the proper phase relationship with the new lighting cycle more rapidly than hepatic bioluminescence did. As previously shown, restricting food access to the daytime altered the phase of hepatic rhythmicity. Our model allowed assessment of the rate of recovery from misalignment once animals were provided with food ad libitum. These studies provide clear evidence for circadian misalignment following environmental perturbations and reveal the utility of this model for minimally invasive, longitudinal monitoring of rhythmicity from specific mouse tissues. |