Evolutionary Systems
Allaby Research Group
Evolutionary systems take into account multiple levels of organization and complex interactions between factors which influence the evolution of an organism. The approach is to produce models which incorporate the influencing factors, and determine which parameters or model structures give rise to data patterns which most closely resemble those observed in reality. In the case of crops, simple phylogenetics will no longer suffice to unravel their evolutionary history: the evolutionary systems approach is required instead. For a synopsis of the argument for this case, see 'Evolutionary systems in crops' below.
The aim in our group is to develop model systems that integrate the following levels of organization:
- the gene level of organization (gene to gene interactions)
- genome architecture (locality and spatial distribution of genes under natural or artificial selection in the genome)
- spatially extended population structure (the locality of the 'whole organism' within an arena where the locality has factors such as hydrology, altitude, temperature)
- interactors causing selection pressures (this could be diseases, or in the example discussed below, the cultivating actions of farmers)
Such models may then be used to establish the patterns of genetic diversity expected under specific (and complex) circumstances, and therefore greatly aid us in how to interpret the genetic diversity we see in the real world. While this approach may be applied to any issue of biodiversity, we explore further our own interests in crop evolution below.
Evolutionary systems in crops
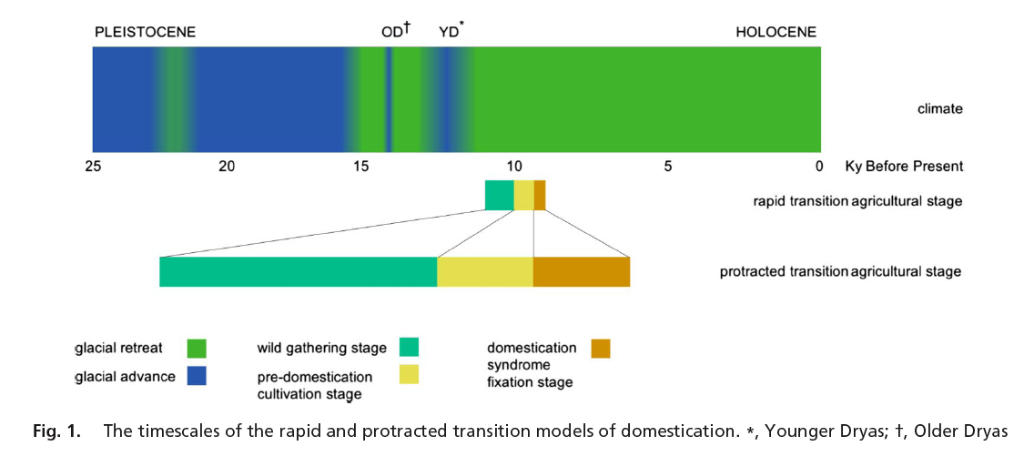
Our understanding of crop origins is undergoing a paradigm shift of major biological significance. Previously, it had been supposed that domesticated crops originated in places such as the Near East around 10,000 years ago in a relatively rapid manner. This would suggest that artificial selection pressure was so strong that natural selection was quickly overcome, and that crop origins were biologically fairly uninteresting, and likely to be discrete geographically. These features are convenient for phylogenetic analyses. However, archaeology has subsequently discovered that the origination of crops was a long drawn out process - the so called protracted transition.
The protracted transition is of major biological significance because artificial selection did not in fact quickly dominate natural selection, rather there was a long interaction between the two processes, mediated through gene flow. Under such conditions one would not expect crops to neatly originate from one area, but for there to have been extensive mixing between regions. However, phylogenetic approaches applied to genome wide data of several botanical species of economic importance suggested a single origin for most crops, seemingly supporting the 'rapid transition' - ie crops associated with discrete geographic localities. We used a population model to explore this anomaly with virtual plants drawn from either one or two highly structured 'wild' populations representing one or two domestication events respectively.
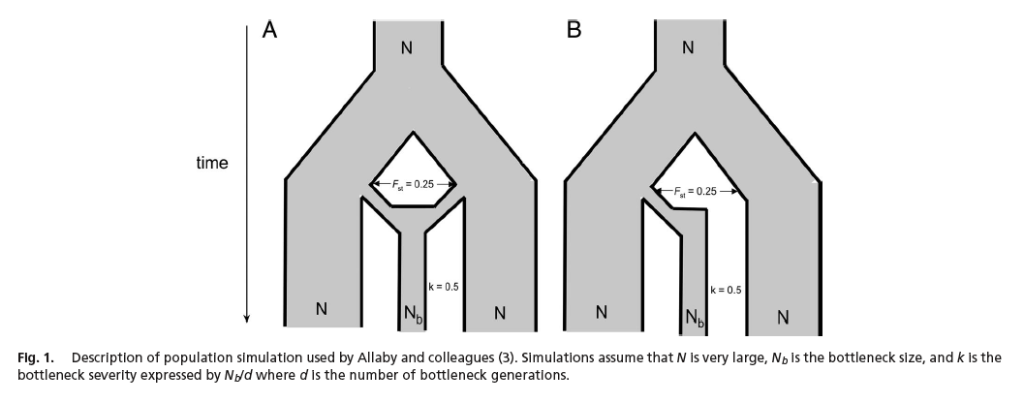
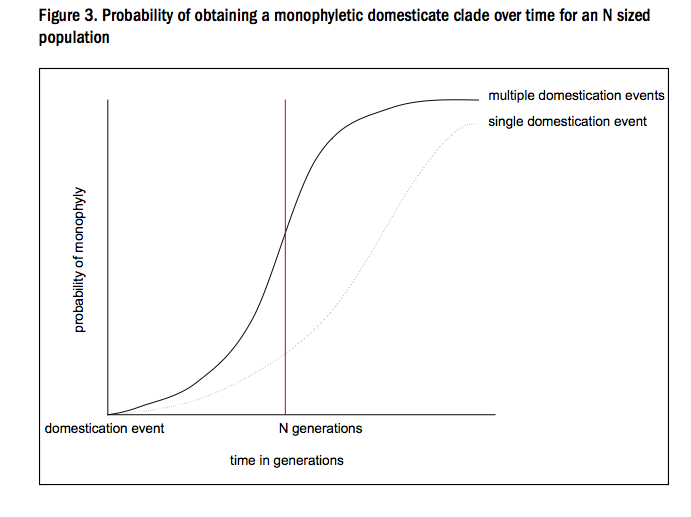
This model showed that not only would both single and multiply domesticated crops produce a signature which appears to be monophyletic, the latter would achieve this more rapidly. Consequently, a protracted transition (multiple origins, gene flow), is actually more likely to look like a single origin than a single origin when phylogenetics are used!
So the anomaly was solved by establishing that the summary statistics being used have little information value. The protracted transition means that the mechanism of the rise of the domestication syndrome (those genes under artificial selection) has become key to understanding crop origins - we need to understand the evolutionary process, because we need to understand how that process shaped genetic diversity. In order to do that we must take into account all of the levels of organization outlined above (genes, genomes, populations, selection pressures) into one system, because they are all interdependent and so have an effect on the resulting diversity. The current aim of our research group is to use computational biology to build such a model system in a modular way to allow general and specific investigation of the effects of these processes on plant genome diversity.
Publications
Allaby RG (2010) Integrating the processes in the evolutionary system of domestication. Journal of Experimental Botany 61:935-944.
Allaby RG, Brown TA, Fuller DQ (2010) A simulation of the effect of inbreeding on crop domestication genetics with comments on the integration of archaeobotany and genetics: a reply to Honne and Heun. Vegetation History and Archaeobotany 19:151-158.
Fuller DQ, Allaby RG, Stevens C (2010) Domestication as Innovation: The entanglement of techniques, technology and chance in the domestication of cereal crops. World Archaeology 42:13-28.
Brown TA, Jones MK, Powell W, Allaby RG (2009) The complex origins of domesticated crops. Trends Ecol. Evol. 24:103-109.
Allaby R.G. (2008) The rise of plant domestication: life in the slow lane. Biologist 55:94-99.
Allaby R.G., Fuller D.Q., Brown T.A. (2008) The genetic expectations of a protracted model for the origins of domesticated crops. Proceedings of the National Academy of Sciences USA 105:13982-13986.
Allaby R.G., Fuller D.Q., Brown T.A. (2008) Reply to Ross-Ibarra & Gaut: Multiple domestications do appear monophyletic if an appropriate model is used. Proceedings of the National Academy of Sciences USA pnas online: doi:10.1073/pnas.0810296105.
Allaby R.G. and Brown T.A. (2004) On the use of trees and crop origins: a reply. Genome 47:621-622.
Allaby R.G. and Brown T.A. (2003) AFLP analysis and the origins of agriculture. Genome 46:448-453.
