Coupled Translation
Coupled translation
Control of gene expression at the level of translation of mRNA is a critical component in the pathway from gene to protein and the eukaryotic translation machinery is able to exploit a number of novel processes to expand the capacity of the system. The evolutionary pressures that have constrained viruses which use RNA as genetic material to maintain small genomes has resulted in the development of strategies to maximise the number of encoded proteins, with many of these being exerted at the translational level. Since viruses depend solely on their host to provide the ribosomal components necessary to synthesise their proteins any translational regulatory mechanism utilised by the virus must be compatible with the host system. Consequently, the study of novel virus translational mechanisms has led to insights into the host molecular processes involved in translation such as capping, function of eukaryotic initiation factors (eIFs) and internal ribosome entry sites (IRES).
Sequence analysis has shown that the M2 mRNAs of all pneumoviruses contain two open reading frames, conserved in location though not in sequence. The first major ORF (ORF-1) of the M2 gene of these viruses utilises 60-75% of the entire coding capacity of the M2 mRNA and the protein product from this ORF, which is readily detected in virus infected cells, is the transcriptional activator. The second major ORF (ORF-2) is located towards the 3’ end of the mRNA, overlaps ORF-1 and utilises the remainder of the unassigned available nucleotide sequence (see figure). The protein products from ORF-2 of the RSV and PVM M2 genes have been detected in virus infected cells and the RSV M2-2 protein has been proposed to be involved in control of the switch between virus RNA replication and transcription.
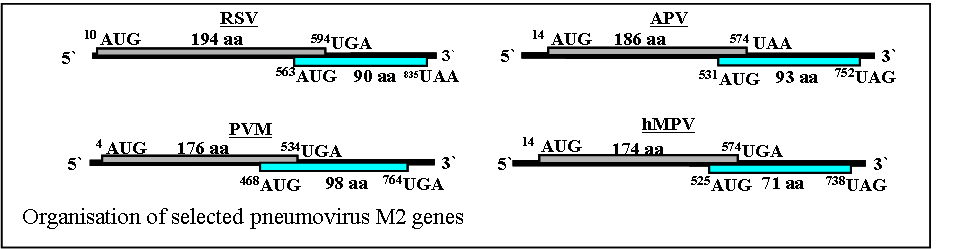
We have shown that ribosomes access the pneumovirus M2 gene second ORF using a novel mechanism in which expression of the M2 ORF-2 protein is initiated at AUGs located upstream of the ORF-1 termination codon, and that expression from these initiation codons requires the prior termination of M2 ORF-1 translation in vivo. Similar coupled translation mechanisms have been described for subgenomic mRNA of the calicivirus rabbit haemorrhagic disease virus (RHDV) which has a 17 nucleotide overlap and in the feline calicivirus which has two ORFs that overlap at the stop and start of ORFs 2 and 3, respectively, with the sequence AUGA.
Surprisingly, the overlap region alone is not sufficient to achieve coupled expression [38]. In the RSV M2 mRNA the coupling process requires at least 148 nts from the 3’ terminal of the M2-1 ORF showing that interactions over a long distance are necessary for the coupled translation to occur. We have also shown that the RSV M2 mRNA itself contains a region of strong secondary structure located close to the position of the sequences required to direct coupled translation.
The M2 mRNAs of PVM and APV also utilise a coupled translation process to access M2 ORF-2. In the PVM M2 mRNA coupled translation is approximately 7-fold less efficient that seen in RSV. However, coupled translation of the second ORF in the APV M2 mRNA is approximately 70 fold lower than the comparable RSV construct.
We are further characterising the coupled translation process to determine the mechanism by which it is achieved.
