Academic Outputs
With over 80% of our research outputs rated as 'internationally excellent' or 'world-leading', the School of Life Sciences continues to drive innovative research that pushes the boundaries of our understanding across the biological sciences.
Here are some of our recent highlights.
View our latest publications and press releases
Emily Skates, Hadrien Delattre, Zoe Schofield, Munehiro Asally and Orkun S. Soyer (2023) Thioflavin T indicates mitochondrial membrane potential in mammalian cells. Biophysical Reports Vol 3, Issue 4
Mitochondrial membrane potential (MMP) is the electrical potential difference across the inner mitochondrial membrane and is essential to the metabolic state of the cell. Our research shows that Thioflavin T (ThT) can act as a MMP indicator in mammalian cells, when used at low concentrations and with low blue-light exposure. However, this dye can also disrupt MMP depending additively on its concentration and blue light exposure. Our findings call for a re-evaluation of ThT’s use at micromolar range in live-cell analyses, but at the same time presents ThT as a potential tool to study MMP dynamics under conditions where TMRM might have limitations (e.g. in studies using microfluidics devices where TMRM can bind to device structure) or where controlled MMP disruption is sought.
Image Credit: Biophysical Reports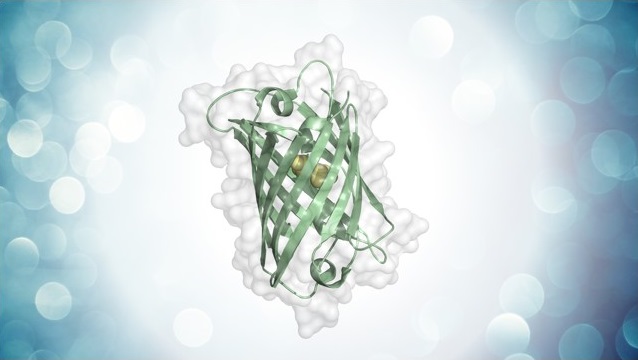
Esther Ivorra-Molla, Dipayan Akhuli, Martin B. L. McAndrew, William Scott, Lokesh Kumar, Saravanan Palani, Masanori Mishima, Allister Crow & Mohan K. Balasubramanian (2023) A monomeric StayGold fluorescent protein. Nature Biotechnology
A collaboration between researchers at the School of Life Sciences and Warwick Medical School has yielded an improved fluorescent protein for use in biological research. mStayGold is highly photo-stable and monomeric version of StayGold that can be used to track the location of proteins inside living cells.
StayGold is an exceptionally bright and stable fluorescent protein that is highly resistant to photobleaching. Despite favorable fluorescence properties, use of StayGold as a fluorescent tag is limited because it forms a natural dimer. Here we report the 1.6 Å structure of StayGold and generate a derivative, mStayGold, that retains the brightness and photostability of the original protein while being fully monomeric.

Beatriz Lagunas, Luke Richards , Chrysi Sergaki, Jamie Burgess, Alonso Javier Pardal, Rana M. F. Hussain, Bethany L. Richmond, Laura Baxter, Proyash Roy, Anastasia Pakidi, Gina Stovold, Saúl Vázquez , Sascha Ott , Patrick Schäfer and Miriam L. Giford (2023) Rhizobial nitrogen fixation efficiency shapes endosphere bacterial communities and Medicago truncatula host growth Microbiome
Despite the knowledge that the soil–plant–microbiome nexus is shaped by interactions amongst its members, very little is known about how individual symbioses regulate this shaping. Even less is known about how the agriculturally important symbiosis of nitrogen-fixing rhizobia with legumes is impacted according to soil type, yet this knowledge is crucial if we are to harness or improve it. In this paper, we asked how the plant, soil and microbiome are modulated by symbiosis between the model legume Medicago truncatula and different strains of rhizobia whose nitrogen-fixing efficiency varies, in three distinct soil types that differ in nutrient fertility, to examine the role of the soil environment upon the plant–microbe interaction during nodulation.
We found that a number of soil edaphic factors including Zn and Mo, and not just the classical N/P/K nutrients, group with microbial community changes, and alterations in the microbiome can be seen across different soil fertility types. The root endosphere emerged as the plant microhabitat more affected by this rhizobial efficiency-driven community reshaping, manifested by the accumulation of members of the phylum Actinobacteria in high-efficient symbiosis systems. The plant in turn plays an active role in regulating its root community, including sanctioning low nitrogen efficiency rhizobial strains, leading to nodule senescence in particular plant–soil–rhizobia strain combinations.
This work opens up the possibility to select inoculation partners best suited for plant, soil type and microbial community to help promote the natural relationship between plants and bacteria, and reduce reliance on environmentally damaging fertilisers, a key sustainability goal as the population grows and crop yields are threatened by climate change.
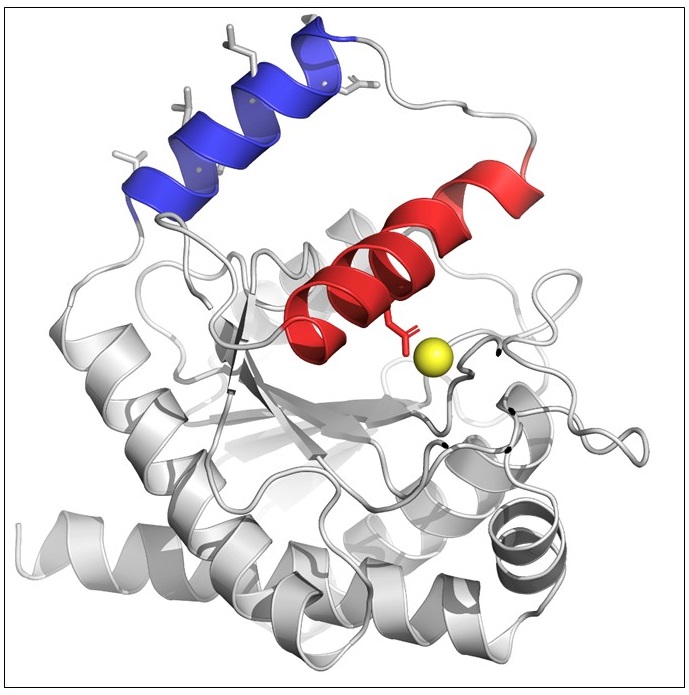
Jonathan Cook, Tyler C Baverstock, Martin BL McAndrew, David I Roper, Phillip J Stansfeld, Allister Crow (2023) Activator-induced conformational changes regulate division-associated peptidoglycan amidases Proceedings of the National Academy of Sciences
The cell envelope defines a bacterium's shape and provides its first defence against antibiotics and the environment.
At the core of the cell envelope is the peptidoglycan layer which provides strength and rigidity to the bacterium and protects it from bursting. The importance of the peptidoglycan layer is readily seen by the large number of antibiotics that work by blocking the layer's construction. These include penicillin, cephalosporin, vancomycin and many others.
When bacteria reproduce, the peptidoglycan layer must be partially broken at the division site to allow the separation of daughter cells. However, the enzymes responsible need to be carefully regulated to avoid self-destruction.
In this paper, our team describes the 3D structure of an enzyme (a peptidoglycan amidase) that is responsible for breaking the peptidoglycan layer and shows how it is regulated by changes in its shape that are triggered by the binding of a partner protein.
The work provides fundamental insights into how bacteria divide and could prompt development of future antimicrobials that either block the enzyme's activity (preventing division) or trigger its uncontrolled activation (causing cells to chew up their own peptidoglycan layer).
Marton, Huba L., Kilbride, Peter, Ahmad, Ashfaq, Sagona, Antonia P. and Gibson, Matthew (2023) Anionic synthetic polymers prevent bacteriophage infection. Journal of the American Chemical Society
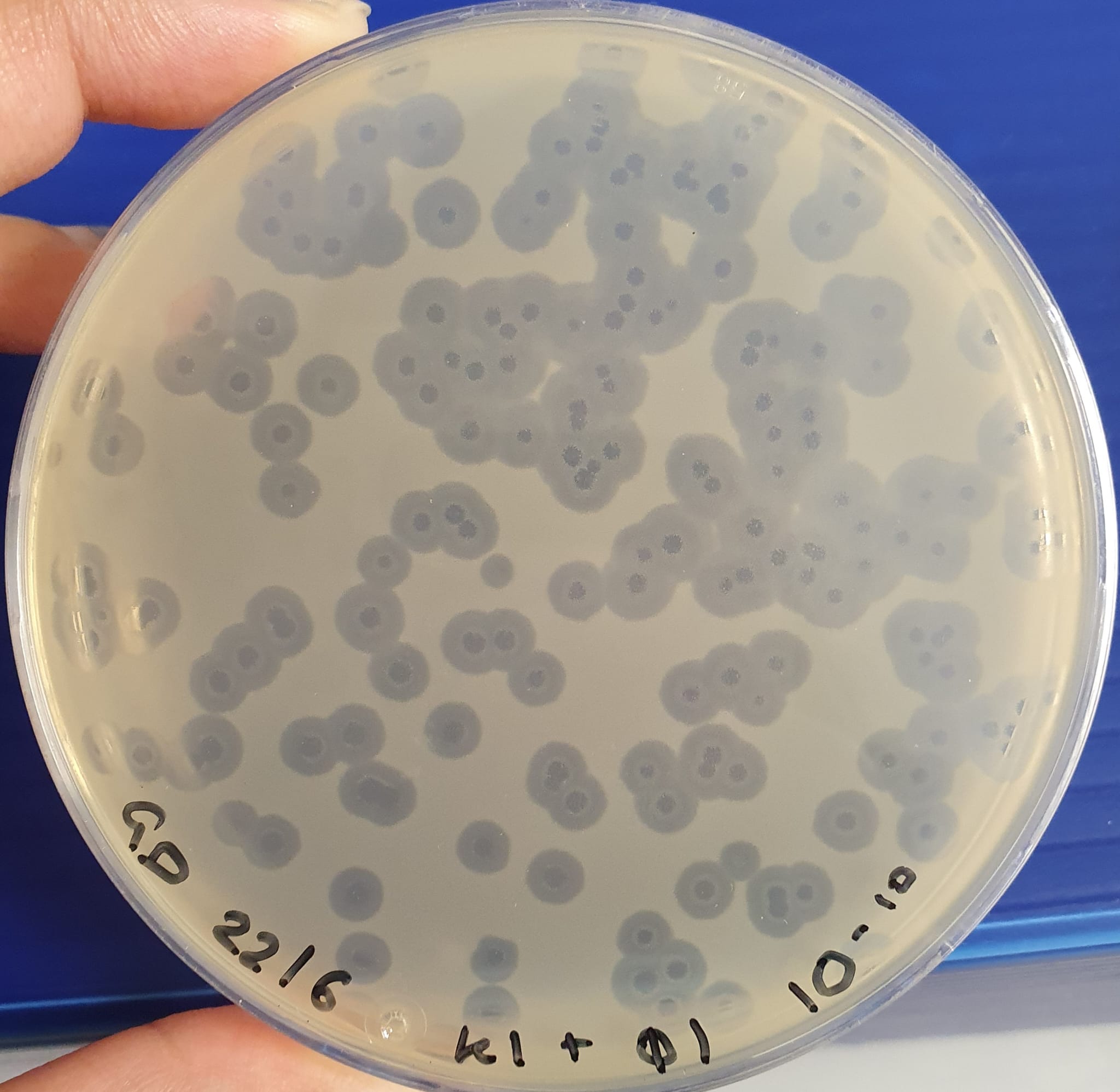
Bioprocessing and biotechnology exploit microorganisms (such as bacteria) for the production of chemicals, biologics, therapies and food. A major unmet challenge is that bacteriophage (phage) contamination compromises products and necessitates shutdowns and extensive decontamination using nonspecific disinfectants. Here, we demonstrate that poly(acrylic acid) prevents phage-induced killing of bacterial hosts, phage replication, while induction of recombinant protein expression is not affected by the presence of the polymer. Poly(acrylic acid) was more active than poly(methacrylic acid) and poly(styrenesulfonate) had no activity showing the importance of the carboxylic acids. Initial evidence supported a virustatic, not virucidal, mechanism of action. This simple, low-cost, mass-produced additive offers a practical, scalable and easy to implement solution to reduce phage contamination.
Image: phage infection as shown by plaque assay
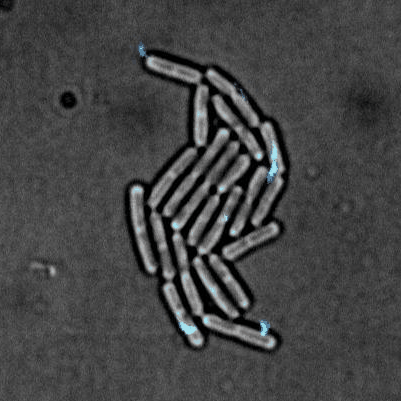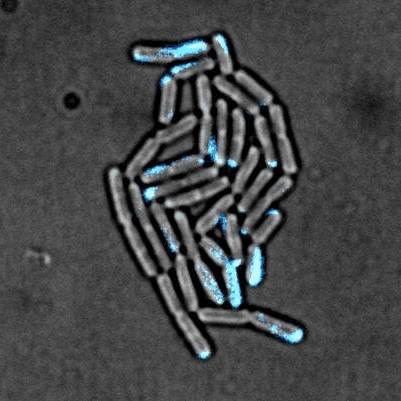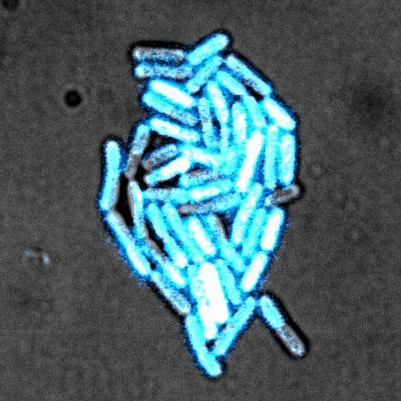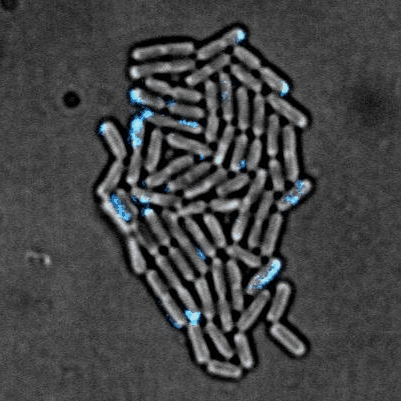
Tailise Carolina de Souza-Guerreiro, Gaia Bondelli, Iago Grobas, Stefano Donini, Valentina Sesti, Chiara Bertarelli, Guglielmo Lanzani, Munehiro Asally & Giuseppe Maria Paternò (2023) Membrane Targeted Azobenzene Drives Optical Modulation of Bacterial Membrane Potential. Advanced Science
Electrical signaling in bacteria plays important roles in biofilm formation, spore formation and germination and antibiotic response. The ability to control cellular membrane potential with precision in space and time could be a new biophysical approach to control bacterial functions. In this study, we show optical modulation of bacterial membrane potential using a photoswitch called ziapin2. Combining fluorescence microscopy, molecular biology and spectroscopy, the results suggested an important role of chloride channel in bacterial electrical signalling. This new optical tool could advance our understanding of bacterial electrophysiology and pave the way for optical control of bacterial cells.
Joint Winner of the Faculty of Science Engineering and Medicine Post-Doctoral Prize 2023 for SLS
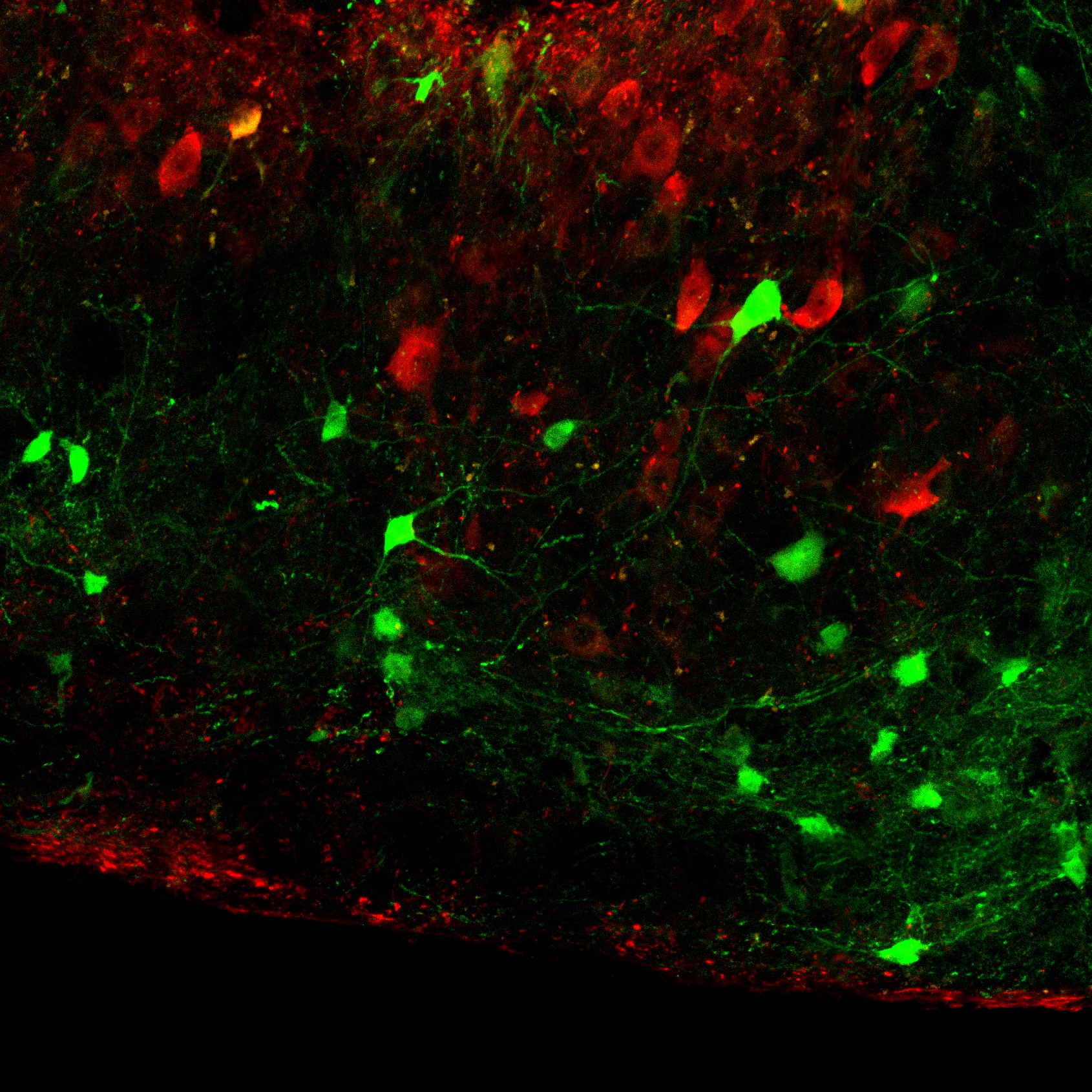
Amol Bhandare, Joseph van de Wiel, Reno Roberts, Ingke Braren, Robert Huckstepp, Nicholas Dale (2022) Analyzing the brainstem circuits for respiratory chemosensitivity in freely moving mice eLIFE
Our paper presents the first recordings and analysis, via head-mounted minicameras, of the activity of brainstem neurons which sense CO2 and control breathing in freely behaving animals. This new technique has enabled us to document a catalogue of neuronal responses to CO2, which show that the detection of CO2 by brainstem neurons is more complex than previously thought. Our work illustrates the feasibility of using imaging, via head mounted minicameras, to analyse the neural mechanisms of breathing and related behaviours such as swallowing and vocalisation in freely behaving animals.
Joint Winner of the Faculty of Science Engineering and Medicine Post-Doctoral Prize 2023 for SLS
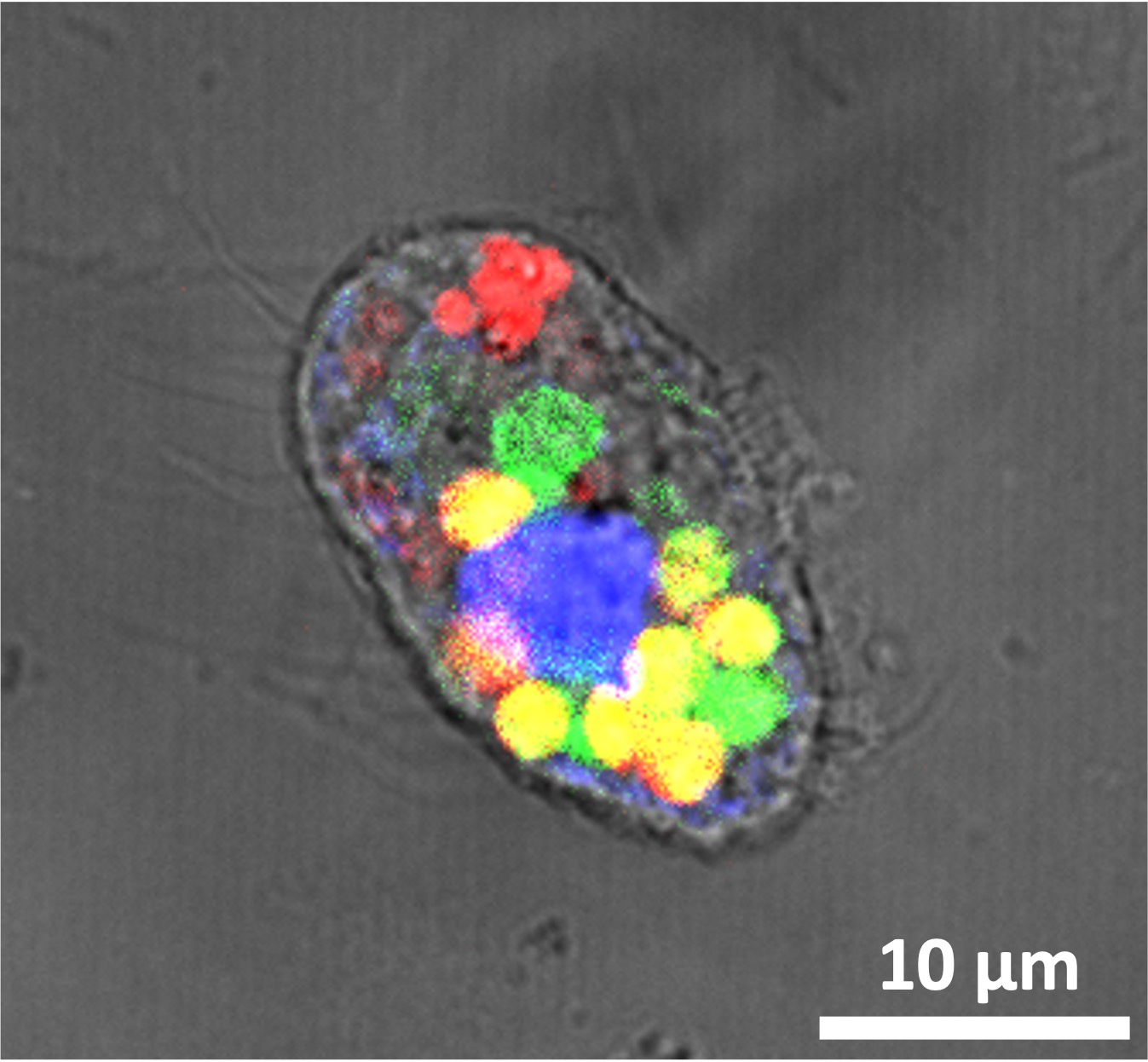
Richard Guillonneau, Andrew R. J. Murphy, Zhao-Jie Teng, Peng Wang, Yu-Zhong Zhang, David J. Scanlan, and Yin Chen (2022) Trade-offs of lipid remodeling in a marine predator–prey interaction in response to phosphorus limitation. PNAS
Microbial growth is often limited by key nutrients like phosphorus. A major response to P limitation is the replacement of membrane phospholipids with non-P lipids. In this study, we uncover a predator–prey interaction trade-off whereby a lipid-remodelled bacterial prey cell becomes more susceptible to grazing by a protozoan predator facilitating its rapid growth. Thus, we highlight a complex interplay between adaptation to the environment and consequences for prey-predator interactions, which may have important implications for the stability and structuring of microbial communities and the performance of the marine food web.
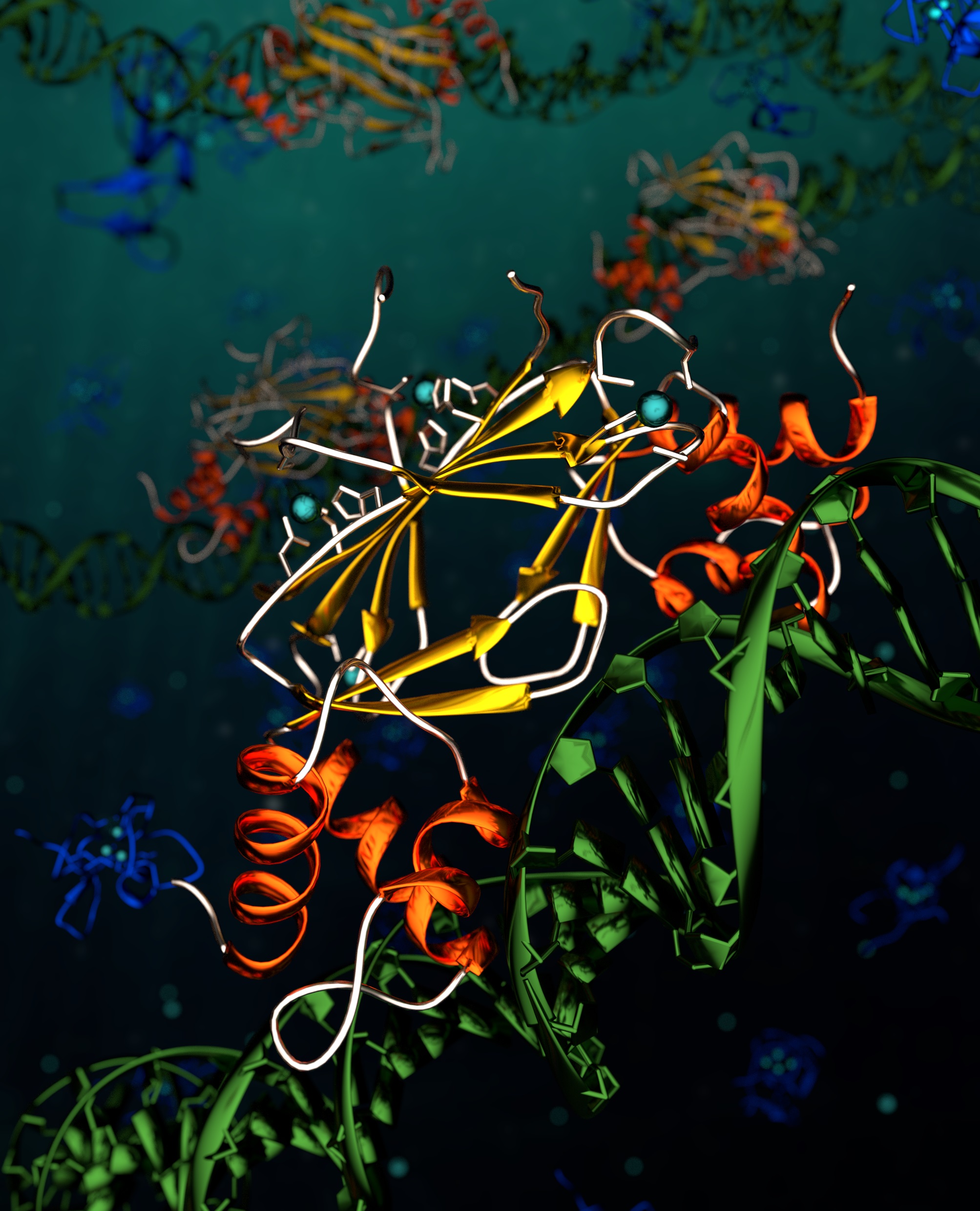
Alevtina Mikhaylina, Amira Z. Ksibe, Rachael C. Wilkinson, Darbi Smith, Eleanor Marks, James P. C. Coverdale, Vilmos Fülöp, David J. Scanlan & Claudia A. Blindauer (2022) A single sensor controls large variations in zinc quotas in a marine cyanobacterium Nature Chemical Biology
The oceans are the somewhat overlooked ‘lungs’ of our planet – every other breath we take is oxygen evolved from systems whilst around a half of the carbon dioxide fixed into biomass on Earth occurs in ocean waters. Marine cyanobacteria are key players in Earth’s ‘lungs’, and this manuscript reveals a novel aspect of their biology, namely the ability to exquisitely regulate zinc homeostasis, a metal often considered to be essential for life. This process allows Synechococcus to vary their internal zinc levels by over a 100-fold and relies on a zinc uptake regulator protein (Zur) which can sense zinc and respond accordingly. This sensor protein also activates a bacterial metallothionein (zinc-binding protein) which, together with highly efficient uptake systems, is responsible for the extraordinary capacity of this organism to accumulate zinc. Given marine cyanobacteria proliferate across nutrient impoverished regions of the words oceans this zinc regulatory feature has undoubtedly contributed to their ability to fulfil this key ‘lungs’ planetary function.
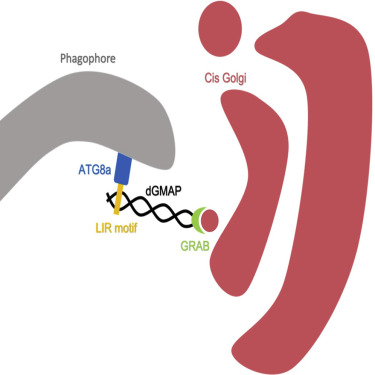
Ashrafur Rahman, Peter Lőrincz, Raksha Gohel, Anikó Nagy, Gábor Csordás, Yan Zhang, Gábor Juhász and Ioannis P. Nezis (2022). GMAP is an Atg8a-interacting protein that regulates Golgi turnover in Drosophila. Cell Reports
View articleThe study of autophagy – the recycling and repair process within cells – has huge potential to aid in fighting the ageing process, bacterial and viral infections and diseases including cancer, Alzheimer’s and Parkinson’s. In this paper, the molecular and cellular mechanisms that regulate selective autophagy in the fruit fly Drosophila melanogaster are identified.
Selective autophagy receptors and adapters contain short linear motifs called LIR motifs (LC3-interacting region), which are required for the interaction with the Atg8-family proteins. LIR motifs bind to the hydrophobic pockets of the LIR motif docking site (LDS) of the respective Atg8-family proteins. The physiological significance of LDS docking sites has not been clarified in vivo. Here, we show that Atg8a-LDS mutant Drosophila flies accumulate autophagy substrates and have reduced lifespan. Using quantitative proteomics to identify the proteins that accumulate in Atg8a-LDS mutants, we identify the cis-Golgi protein GMAP (Golgi microtubule-associated protein) as a LIR motif-containing protein that interacts with Atg8a. GMAP LIR mutant flies exhibit accumulation of Golgi markers and elongated Golgi morphology. Our data suggest that GMAP mediates the turnover of Golgi by selective autophagy to regulate its morphology and size via its LIR motif-mediated interaction with Atg8a.
Understanding the molecular mechanisms of selective autophagy of Golgi complex in cells will help open new avenues of research that will assist elucidating the underlying cellular mechanisms of diseases.
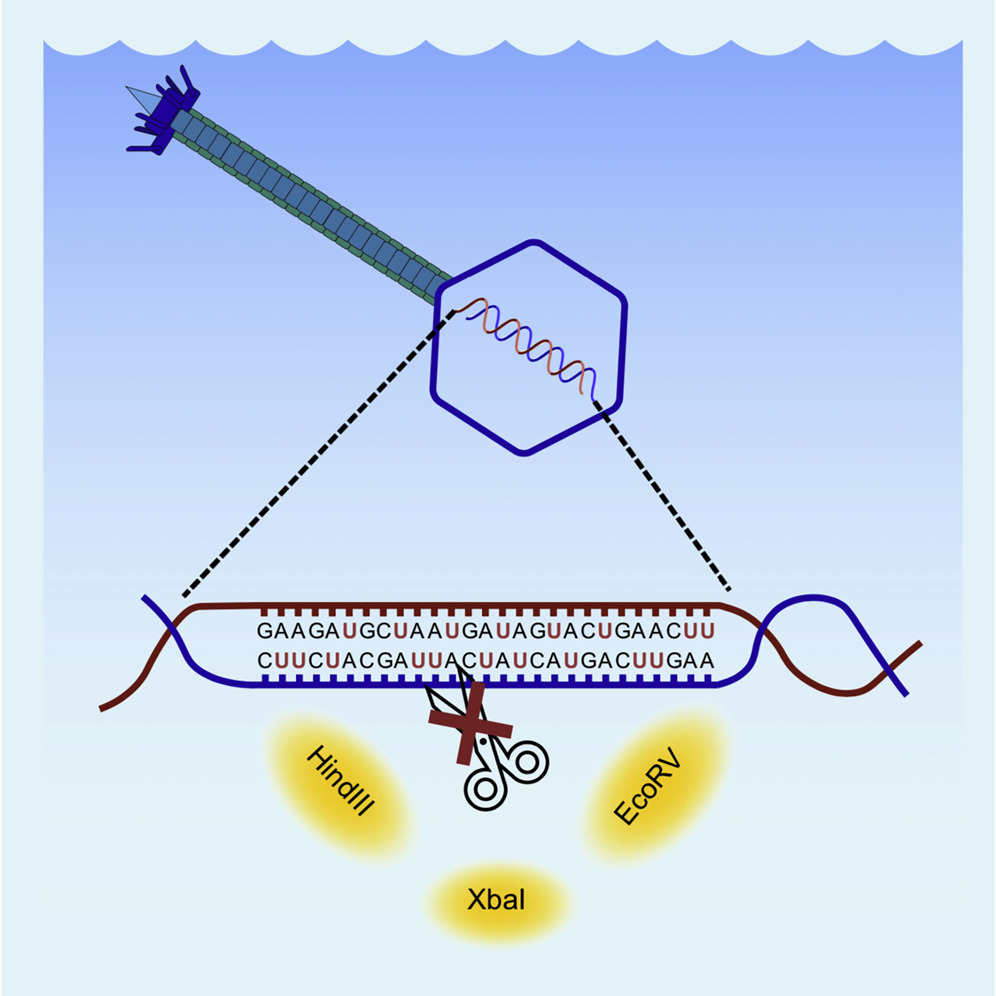
Branko Rihtman, Richard J. Puxty, Alexia Hapeshi, Yan-Jiun Lee, Yuanchao Zhan, Slawomir Michniewski, Nicholas R. Waterfield, Feng Chen, Peter Weigele, Andrew D. Millard,David J. Scanlan, and Yin Chen. (2021) A new family of globally distributed lytic roseophages with unusual deoxythymidine to deoxyuridine substitution. Current Biology.
Marine bacterial viruses (bacteriophages) are abundant biological entities that are vital for shaping microbial diversity, impacting marine ecosystem function, and driving host evolution. The marine roseobacter clade (MRC) is a ubiquitous group of heterotrophic bacteria that are important in the elemental cycling of various nitrogen, sulfur, carbon, and phosphorus compounds. Bacteriophages infecting MRC (roseophages) have thus attracted much attention and more than 30 roseophages have been isolated the majority of which belong to the N4-like group (Podoviridae family) or the Chi-like group (Siphoviridae family), although ssDNA-containing roseophages are also known. In our attempts to isolate lytic roseophages, we obtained two new phages (DSS3_VP1 and DSS3_PM1) infecting the model MRC strain Ruegeria pomeroyi DSS-3. Here, we show that not only do these phages have unusual substitution of deoxythymidine with deoxyuridine (dU) in their DNA, but they are also phylogenetically distinct from any currently known double-stranded DNA bacteriophages, supporting the establishment of a novel family (“Naomiviridae”). These dU-containing phages possess DNA that is resistant to the commonly used library preparation method for metagenome sequencing, which may have caused significant underestimation of their presence in the environment. Nevertheless, our analysis of Tara Ocean metagenome datasets suggests that these unusual bacteriophages are of global importance and more diverse than other well-known bacteriophages, e.g., the Podoviridae in the oceans, pointing to an overlooked role for these novel phages in the environment.
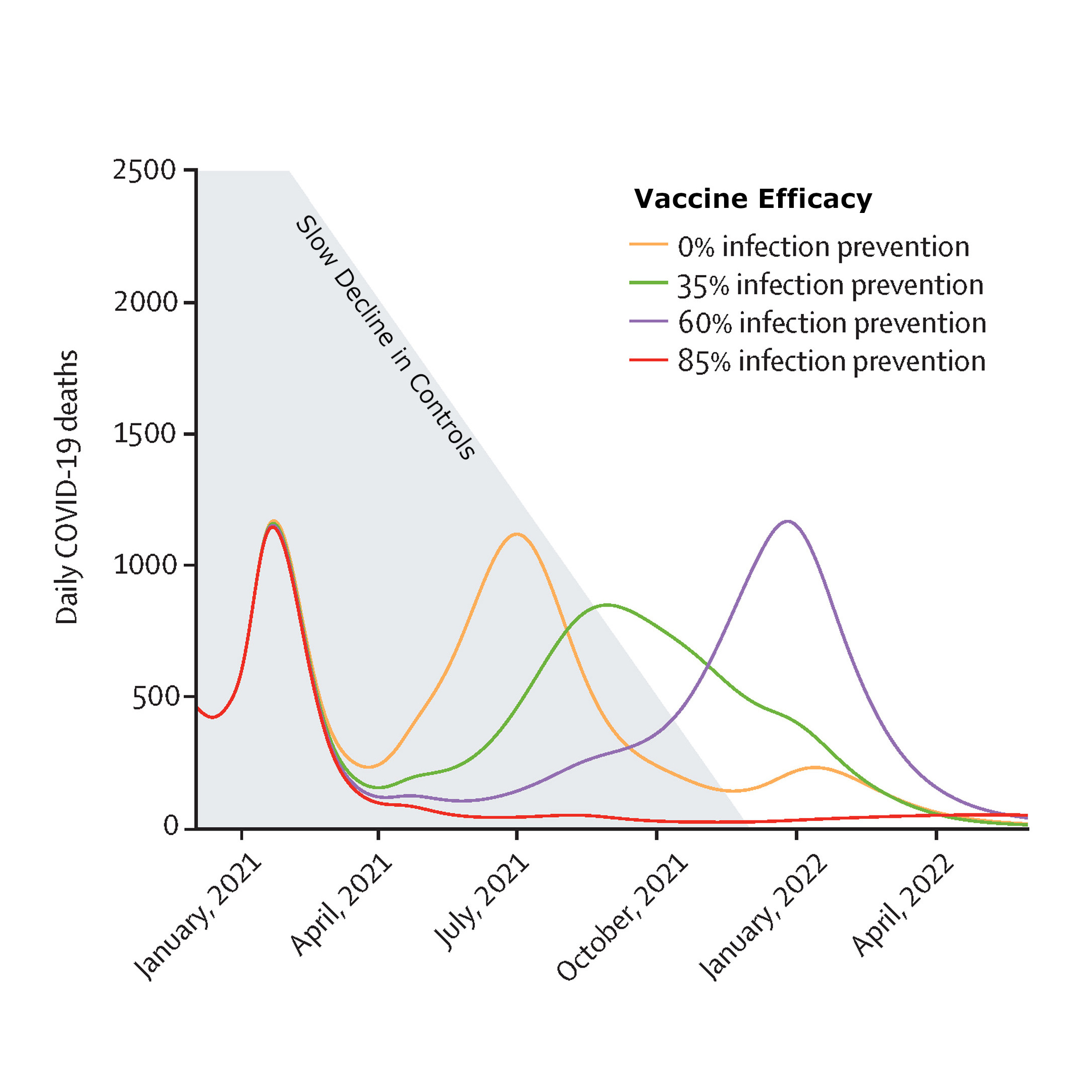
Moore, Sam, Hill, Edward M., Tildesley, Michael J., Dyson, Louise and Keeling, Matt J. (2021) Vaccination and non-pharmaceutical interventions for COVID-19: a mathematical modelling study. Lancet Infectious Diseases.
By combining models of vaccination with the methods of forwards projection, we considered the interaction between the relaxation of non-pharamaceutical interventions (NPIs) and the protection offered by the vaccine. This paper set the tone for unlocking the UK in 2021: Our modelled scenarios highlighted the risks associated with early or rapid relaxation of NPIs, stressing the need for slow release of control measures if large-scale waves of infection are to be avoided. We conclude that while the vaccines against SARS-CoV-2 offer a potential exit strategy for the pandemic, success is highly contingent on the precise vaccine properties and population uptake.
Shanshan Zhou, Hussain Bhukya, Nicolas Malet, Peter J. Harrison, Dean Rea, Matthew J. Belousoff, Hariprasad Venugopal, Paulina K. Sydor, Kathryn M. Styles, Lijiang Song, Max J. Cryle, Lona M. Alkhalaf, Vilmos Fülöp, Gregory L. Challis and Christophe Corre. (2021) Molecular basis for control of antibiotic production by a bacterial hormone. Nature.
Actinobacteria produce numerous antibiotics and other specialized metabolites that have important applications in medicine and agriculture1. Diffusible hormones frequently control the production of such metabolites by binding TetR family transcriptional repressors (TFTRs), but the molecular basis for this remains unclear2. The production of methylenomycin antibiotics in Streptomyces coelicolor A3(2) is initiated by the binding of 2-alkyl-4-hydroxymethylfuran-3-carboxylic acid (AHFCA) hormones to the TFTR MmfR3. Here we report the X-ray crystal structure of an MmfR–AHFCA complex, establishing the structural basis for hormone recognition. We also elucidate the mechanism for DNA release upon hormone binding through the single-particle cryo-electron microscopy structure of an MmfR–operator complex. DNA binding and release assays with MmfR mutants and synthetic AHFCA analogues define the role of individual amino acid residues and hormone functional groups in ligand recognition and DNA release. These findings will facilitate the exploitation of actinobacterial hormones and their associated TFTRs in synthetic biology and in the discovery of new antibiotics.

Sally Hilton, Emma Picot, Susanne Schreiter, David Bass, Keith Norman, Anna E. Oliver, Jonathan D. Moore, Tim H. Mauchline, Peter R. Mills, Graham R. Teakle, Ian M. Clark, Penny R. Hirsch, Christopher J. van der Gast and Gary D. Bending. (2021) Identification of microbial signatures linked to oilseed rape yield decline at the landscape scale. Microbiome.
The plant microbiome plays a vital role in determining host health and productivity. However, we lack real-world comparative understanding of the factors which shape assembly of its diverse biota, and crucially relationships between microbiota composition and plant health. Here we investigated landscape scale rhizosphere microbial assembly processes in oilseed rape (OSR), the UK’s third most cultivated crop by area and the world's third largest source of vegetable oil, which suffers from yield decline associated with the frequency it is grown in rotations. We show that at the landscape scale, OSR crop yield is governed by interplay between complex communities of both pathogens and beneficial biota which is modulated by rotation frequency. Our comprehensive study has identified signatures of dysbiosis within the OSR microbiome, grown in real-world agricultural systems, which could be used in strategies to promote crop yield.
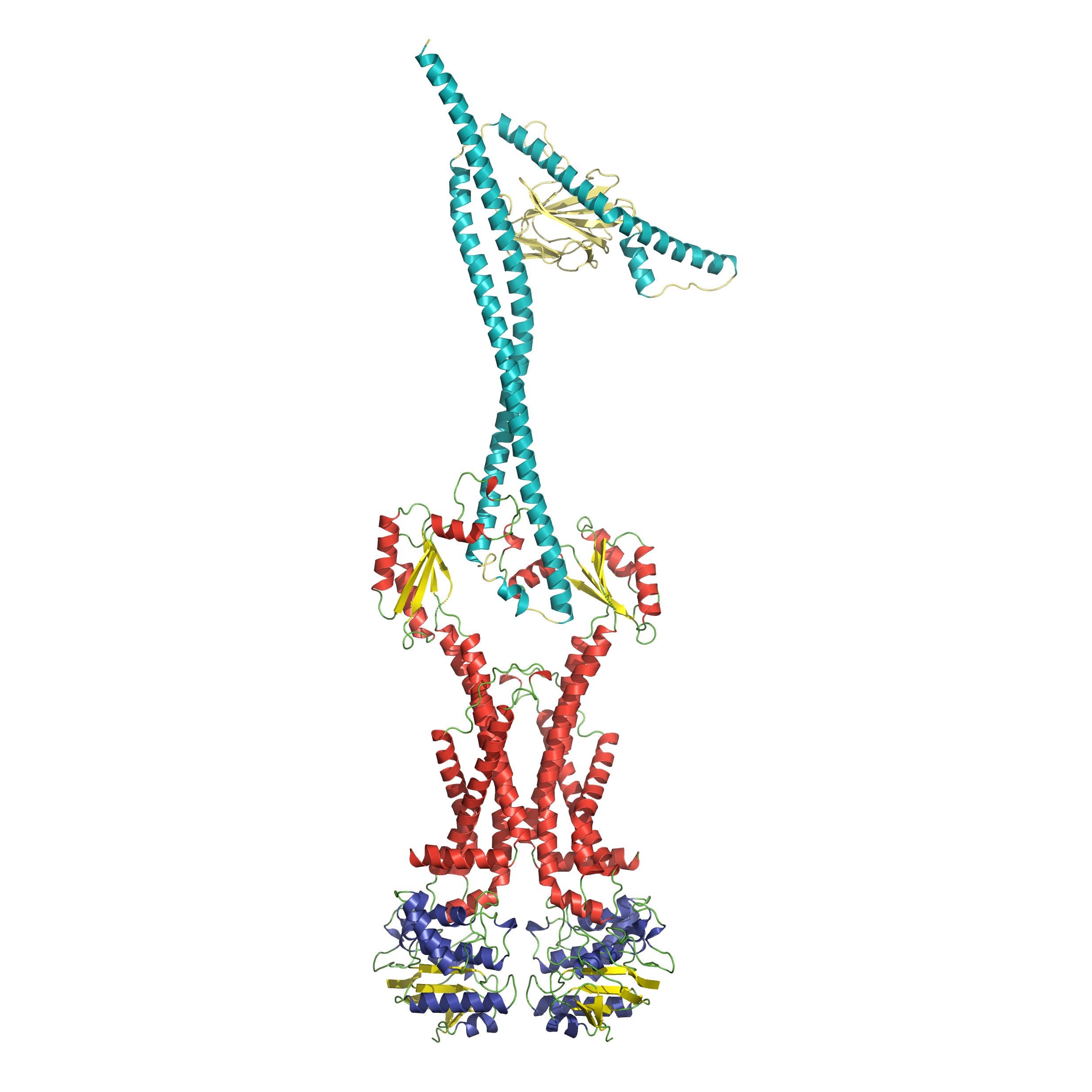
Jonathan Cook, Tyler C. Baverstock, Martin B.L. McAndrew, Phillip J. Stansfeld, David I. Roper, Allister Crow. (2020) Insights into bacterial cell division from a structure of EnvC bound to the FtsX periplasmic domain. PNAS.
The peptidoglycan layer is a core component of the bacterial cell envelope that provides a barrier to the environment and protection from osmotic shock. During division, bacteria must break and rebuild the peptidoglycan layer to enable separation of daughter cells. In E. coli, two of the three amidases responsible (AmiA and AmiB) are regulated by a single periplasmic activator (EnvC) that is, itself, controlled by an atypical ABC transporter (FtsEX) tethered to the cytoplasmic septal Z-ring. Here we define the structural basis for the interaction of FtsEX with EnvC and suggest a molecular mechanism for amidase activation where EnvC autoinhibition is relieved by ATP-driven conformational changes transmitted through the FtsEX-EnvC complex.
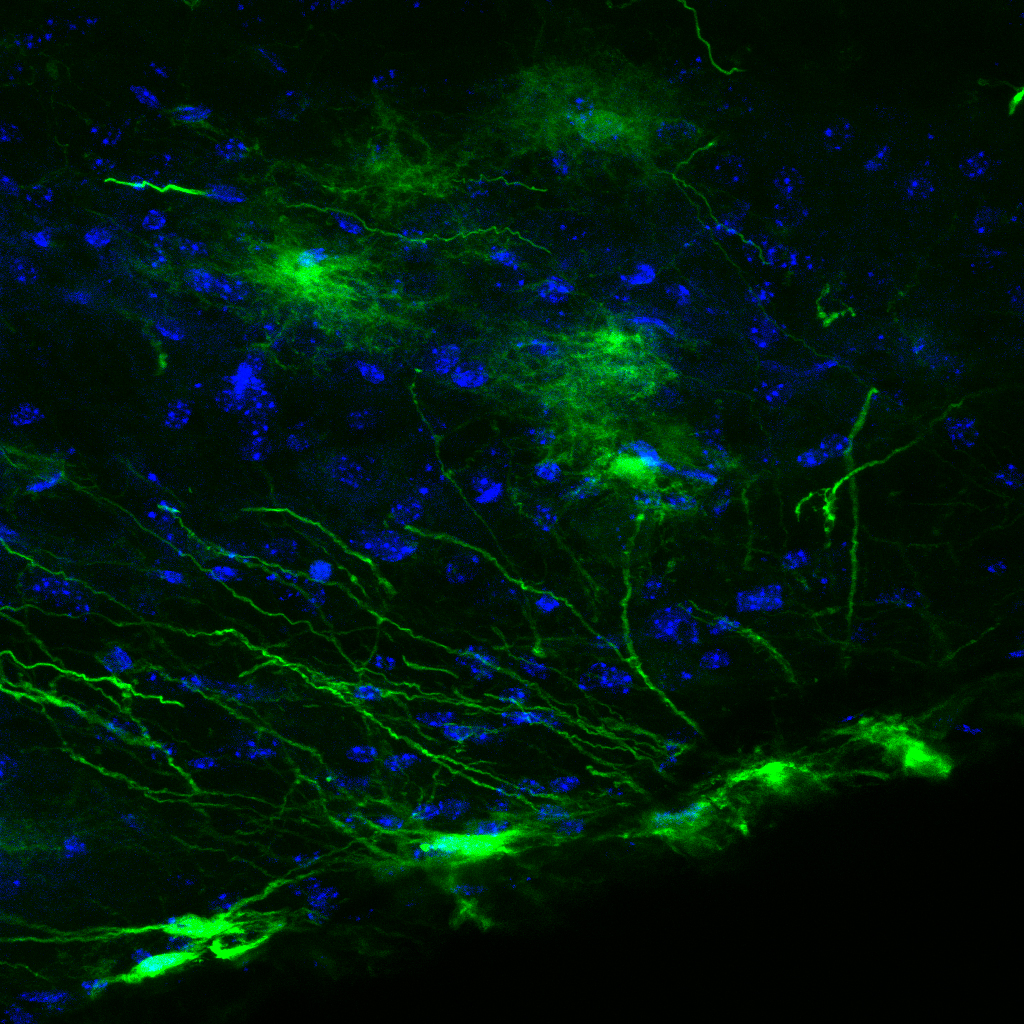
Jan de Wiel J, Meigh L, Bhandare A, Cook J, Nijjar S, Huckstepp RT & Dale N. (2020) Connexin26 mediates CO2-dependent regulation of breathing via glial cells of the medulla oblongata. Communications Biology.
Breathing is highly sensitive to the PCO2 of arterial blood. Although CO2 is detected via the proxy of pH, CO2 acting directly via Cx26 may also contribute to the regulation of breathing. Here we exploit our knowledge of the structural motif of CO2-binding to Cx26 to devise a dominant negative subunit (Cx26DN) that removes the CO2-sensitivity from endogenously expressed wild type Cx26. Expression of Cx26DN in glial cells of a circumscribed region of the mouse medulla - the caudal parapyramidal area – reduced the adaptive change in tidal volume and minute ventilation by approximately 30% at 6% inspired CO2. As central chemosensors mediate about 70% of the total response to hypercapnia, CO2-sensing via Cx26 in the caudal parapyramidal area contributed about 45% of the centrally-mediated ventilatory response to CO2. Our data unequivocally link the direct sensing of CO2 to the chemosensory control of breathing and demonstrates that CO2-binding to Cx26 is a key transduction step in this fundamental process.

Tildesley, MJ, Brand, S, Brooks Pollock, E, Bradbury, MV, Werkman, M & Keeling, MJ. (2019) The role of movement restrictions in limiting the economic impact of livestock infections. Nature Sustainability.
Livestock movements are essential for the economic success of the industry. However, these movements come with the risk of long-range spread of infection, potentially bringing infection to previously disease-free areas where subsequent localized transmission can be devastating. Mechanistic predictive models usually consider controls that minimize the number of livestock affected without considering other costs of an ongoing epidemic. However, it is more appropriate to consider the economic burden, as movement restrictions have major consequences for the economic revenue of farms. Here, using mechanistic models of foot-and-mouth disease, bluetongue virus and bovine tuberculosis in the UK, we compare the economically optimal control strategies for these diseases. We show that for foot-and-mouth disease, the optimal strategy is to ban movements in a small radius around infected farms; the balance between disease control and maintaining ‘business as usual’ varies between regions. For bluetongue virus and bovine tuberculosis, we find that the cost of any movement ban is greater than the epidemiological benefits due to the low within-farm prevalence and slow rate of disease spread. This work suggests that movement controls need to be carefully matched to the epidemiological and economic consequences of the disease, and that optimal movement bans are often of far shorter duration than allowed under existing policy.

Smith O, Nicholson WV, Kistler L, Mace E, Clapham A, Rose P, Stevens C, Ware R, Samavedam S, Barker G, Jordan D, Fuller DQ, Allaby RG. (2019) A domestication history of dynamic adaptation and genomic deterioration in Sorghum. Nature Plants.
The evolution of domesticated cereals was a complex interaction of shifting selection pressures and repeated episodes of introgression. Genomes of archaeological crops have the potential to reveal these dynamics without being obscured by recent breeding or introgression. We report a temporal series of archaeogenomes of the crop sorghum (Sorghum bicolor) from a single locality in Egyptian Nubia. These data indicate no evidence for the effects of a domestication bottleneck, but instead reveal a steady decline in genetic diversity over time coupled with an accumulating mutation load. Dynamic selection pressures acted sequentially to shape architectural and nutritional domestication traits and to facilitate adaptation to the local environment. Later introgression between sorghum races allowed the exchange of adaptive traits and achieved mutual genomic rescue through an ameliorated mutation load. These results reveal a model of domestication in which genomic adaptation and deterioration were not focused on the initial stages of domestication but occurred throughout the history of cultivation.
