Population Modelling
Allaby Research Group
Models are extremely useful tools to address questions that are too complex to describe formally. At ARG we develop software to address various questions about populations to help us interpret the data we see in the 'real world'. One example of model use is through the hypothetical crop models we apply to help track the fate of biomarker types (Allaby and Brown 2003, Allaby, Merriwether and Brown 2007, submitted), described in more detail below.
As the example below illustrates, models help us to establish what we should expect to see, given certain processes. All too often, what we think we should expect, we should not.
Designing and running models of crop domestication and general population processes is an ongoing research area at ARG.

Wild hypothetical crop
Domesticated hypothetical crop
In this model we ascribe virtual plants with chromosomes, each with a set of biallelic markers. We then subject the crops to various domestication scenarios such as multiple and single domestication events. In this case we find that biallelic markers such as AFLPs are actually more likely to produce monophyletic groups under the scenario of multiple domestications. This is important because many studies have inferred single domestication events, and localities, on the basis of phylogenetic analyses of AFLP data giving rise to monophyletic branches for domesticate crops.
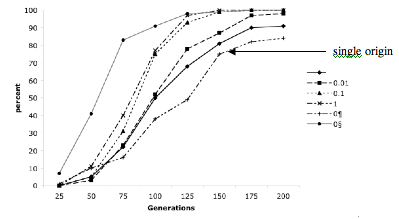
This figure shows 4800 trials (each point represents 100 trials) in which a subset of a wild population or populations was brought into cultivation, after a time independently domesticated groups were allowed to amalgamate, the populations were then cycled for between 25 and 200 generations (X-axis). Samples of wild forms and domesticated forms were then drawn, and a phylogenetic tree calculated. The Y-axis displays the number of times out of 100 trials that the domesticated group was monophyletic. This particular study demonstrates that single domestications (ie no amalgamation) are actually slowest to reach monophyly. While this may appear to be counterintuitive, it actually makes a lot of sense when you consider single domestication populations only have drift to differentiate them from their wild progenitors, whereas multiple domesticate populations have both drift and the influx of alleles and frequencies unrelated to each wild progenitor population.
Modelling publications
Allaby R.G., Fuller D.Q., Brown T.A. (2008) The genetic expectations of a protracted model for the origins of domesticated crops. Proceedings of the National Academy of Sciences USA 105:13982-13986.
Allaby R.G. and Brown T.A. (2004) On the use of trees and crop origins: a reply. Genome 47:621-622.
Allaby R.G. and Brown T.A. (2003) AFLP analysis and the origins of agriculture. Genome 46:448-453.
