Molecular Control of Flowering and Development

We are interested in the molecular basis of how flowering is controlled in plants (both Arabidopsis and crop plants), and one of our interests is in the control of flowering by daylength (photoperiod). The Arabidopsis day neutral flowering (dnf) mutant is one of the mutants that we have isolated and characterised. We showed that the DNF gene encodes an E3 ligase that regulates the expression of the CONSTANS gene and thus flowering time.
CRISPR gene editing
We are using the revolutionary CRISPR gene-editing technology to gene-edit endogenous flowering genes in plants to enable us to manipulate their flowering time.
We have done this in Arabidopsis, creating mutations in the endogenous FT gene which results in late flowering plants (eg. FT1_4, FT1_5 & FT1_K shown in the figure below). The FT gene in these plant lines has been modified by the CRISPR/Cas9 and has had an Adenine residue either inserted or deleted 3 nucleotides upstream of the PAM site, thus resulting in inactivation of the FT gene and late flowering of those plants.
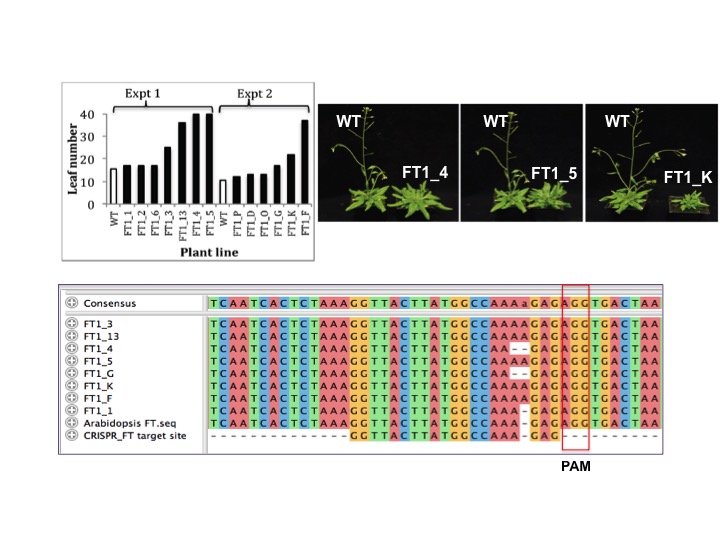
More recently our research is focussed on delivering CRISPR/Cas9 into plants using RNA virus expression vectors (see our Virus induced flowering page) in order to avoid the need to make transgenic plants, so the resulting gene-edited plants will have no additional heterologous DNA inserted in them.
Recent publications

