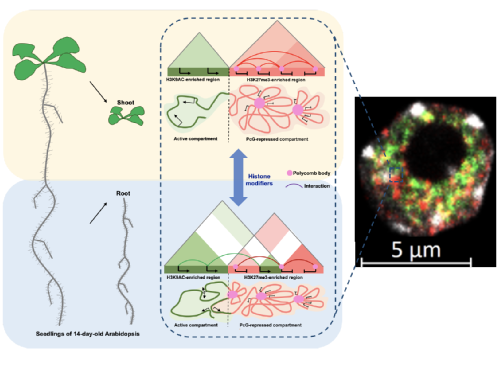
Chromatin dynamics and developmental reprogramming
Histone modifications play a critical role in the control of growth, development, and adaptation to environmental fluctuations of most multicellular eukaryotes. We are interested in the role that histone methylases and demethylases play in chromatin dynamics and developmental reprogramming in plants. We have found that failure to reset chromatin marks during sexual reproduction results in the transgenerational inheritance of histone marks, which causes transposon activation and alters the genome of the offspring. We have also found that these histone modifying enzymes are critical to regulate 3D genome organization and ultimately shape the transcriptional reprogramming underpining plant development.
See our latest publications:
Antunez-Sanchez, J., et al., (2020) A new role for histone demethylases in the maintenance of plant genome integrity. eLife doi:10.1101/2020.03.02.972752
Huang, Y., et al., (2021) Polycomb-dependent differential chromatin compartmentalization determines gene co-regulation in Arabidopsis. Genome Research (doi:10.1101/gr.273771.120).